Regulation of phosphate homeostasis by MicroRNA in Arabidopsis
- PMID: 16387831
- PMCID: PMC1356548
- DOI: 10.1105/tpc.105.038943
Regulation of phosphate homeostasis by MicroRNA in Arabidopsis
Abstract
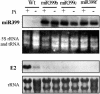
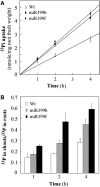
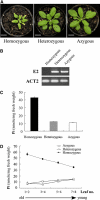
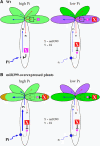
In this study, we reveal a mechanism by which plants regulate inorganic phosphate (Pi) homeostasis to adapt to environmental changes in Pi availability. This mechanism involves the suppression of a ubiquitin-conjugating E2 enzyme by a specific microRNA, miR399. Upon Pi starvation, the miR399 is upregulated and its target gene, a ubiquitin-conjugating E2 enzyme, is downregulated in Arabidopsis thaliana. Accumulation of the E2 transcript is suppressed in transgenic Arabidopsis overexpressing miR399. Transgenic plants accumulated five to six times the normal Pi level in shoots and displayed Pi toxicity symptoms that were phenocopied by a loss-of-function E2 mutant. Pi toxicity was caused by increased Pi uptake and by translocation of Pi from roots to shoots and retention of Pi in the shoots. Moreover, unlike wild-type plants, in which Pi in old leaves was readily retranslocated to other developing young tissues, remobilization of Pi in miR399-overexpressing plants was impaired. These results provide evidence that miRNA controls Pi homeostasis by regulating the expression of a component of the proteolysis machinery in plants.
Figures




References
-
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. - PubMed
-
- Ames, B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8 115–118.
-
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6 463–470. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

