Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica
- PMID: 16389302
- PMCID: PMC1323478
- DOI: 10.1371/journal.ppat.0010045
Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica
Abstract
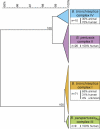
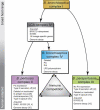
Bordetella pertussis, B. bronchiseptica, B. parapertussis(hu), and B. parapertussis(ov) are closely related respiratory pathogens that infect mammalian species. B. pertussis and B. parapertussis(hu) are exclusively human pathogens and cause whooping cough, or pertussis, a disease that has resurged despite vaccination. Although it most often infects animals, infrequently B. bronchiseptica is isolated from humans, and these infections are thought to be zoonotic. B. pertussis and B. parapertussis(hu) are assumed to have evolved from a B. bronchiseptica-like ancestor independently. To determine the phylogenetic relationships among these species, housekeeping and virulence genes were sequenced, comparative genomic hybridizations were performed using DNA microarrays, and the distribution of insertion sequence elements was determined, using a collection of 132 strains. This multifaceted approach distinguished four complexes, representing B. pertussis, B. parapertussis(hu), and two distinct B. bronchiseptica subpopulations, designated complexes I and IV. Of the two B. bronchiseptica complexes, complex IV was more closely related to B. pertussis. Of interest, while only 32% of the complex I strains were isolated from humans, 80% of the complex IV strains were human isolates. Comparative genomic hybridization analysis identified the absence of the pertussis toxin locus and dermonecrotic toxin gene, as well as a polymorphic lipopolysaccharide biosynthesis locus, as associated with adaptation of complex IV strains to the human host. Lipopolysaccharide structural diversity among these strains was confirmed by gel electrophoresis. Thus, complex IV strains may comprise a human-associated lineage of B. bronchiseptica from which B. pertussis evolved. These findings will facilitate the study of pathogen host-adaptation. Our results shed light on the origins of the disease pertussis and suggest that the association of B. pertussis with humans may be more ancient than previously assumed.
Conflict of interest statement
Figures



References
-
- van der Zee A, Groenendijk H, Peeters M, Mooi FR. The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int J Syst Bacteriol. 1996;46:640–647. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

