Mode of action and subsite studies of the guluronan block-forming mannuronan C-5 epimerases AlgE1 and AlgE6
- PMID: 16390328
- PMCID: PMC1422759
- DOI: 10.1042/BJ20051804
Mode of action and subsite studies of the guluronan block-forming mannuronan C-5 epimerases AlgE1 and AlgE6
Abstract
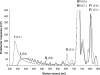
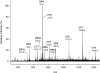
AlgE1, AlgE5 and AlgE6 are members of a family of mannuronan C-5 epimerases encoded by the bacterium Azotobacter vinelandii, and are active in the biosynthesis of alginate, where they catalyse the post-polymerization conversion of beta-D-mannuronic acid (M) residues into alpha-L-guluronic acid residues (G). All enzymes show preference for introducing G-residues neighbouring a pre-existing G. They also have the capacity to convert single M residues flanked by G, thus 'condensing' G-blocks to form almost homopolymeric guluronan. Analysis of the length and distribution of G-blocks based on specific enzyme degradation combined with size-exclusion chromatography, electrospray ionization MS, HPAEC-PAD (high-performance anion-exchange chromatography and pulsed amperometric detection), MALDI (matrix-assisted laser-desorption ionization)-MS and NMR revealed large differences in block length and distribution generated by AlgE1 and AlgE6, probably reflecting their different degree of processivity. When acting on polyMG as substrates, AlgE1 initially forms only long homopolymeric G-blocks >50, while AlgE6 gives shorter blocks with a broader block size distribution. Analyses of the AlgE1 and AlgE6 subsite specificities by the same methodology showed that a mannuronan octamer and heptamer respectively were the minimum substrate chain lengths needed to accommodate enzyme activities. The fourth M residue from the non-reducing end is epimerized first by both enzymes. When acting on MG-oligomers, AlgE1 needed a decamer while AlgE6 an octamer to accommodate activity. By performing FIA (flow injection analysis)-MS on the lyase digests of epimerized and standard MG-oligomers, the M residue in position 5 from the non-reducing end was preferentially attacked by both enzymes, creating an MGMGGG-sequence (underlined and boldface indicate the epimerized residue).
Figures





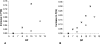

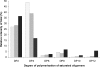
References
-
- Painter T. J. Algal polysaccharides. In: Aspinall G. O., editor. The Polysaccharides, vol. 2. New York: Academic Press; 1983. pp. 195–285.
-
- Gorin P. A. J., Spencer J. F. T. Exocellular alginic acid from Azotobacter vinelandii. Can. J. Chem. 1966;44:993–998.
-
- Govan J. R. W., Fyfe J. A. M., Jarman T. R. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J. Gen. Microbiol. 1981;125:217–220. - PubMed
-
- Ertesvåg H., Høidal H. K., Hals I. K., Rian A., Doseth B., Valla S. A family of modular type mannuronan C-5 epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 1995;16:719–731. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

