Use of quantitative real-time PCR to study the kinetics of extracellular DNA released from Candida albicans, with implications for diagnosis of invasive Candidiasis
- PMID: 16390962
- PMCID: PMC1351963
- DOI: 10.1128/JCM.44.1.143-150.2006
Use of quantitative real-time PCR to study the kinetics of extracellular DNA released from Candida albicans, with implications for diagnosis of invasive Candidiasis
Abstract
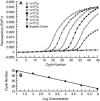
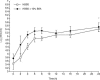
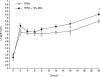
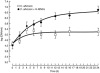
Quantitative real-time PCR (qPCR) is considered one of the most sensitive methods to detect low levels of DNA from pathogens in clinical samples. To improve the design of qPCR for the detection of deeply invasive candidiasis, we sought to develop a more comprehensive understanding of the kinetics of DNA released from Candida albicans in vitro and in vivo. We developed a C. albicans-specific assay targeting the rRNA gene complex and studied the kinetics of DNA released from C. albicans alone, in the presence of human blood monocytes (H-MNCs), and in the bloodstream of rabbits with experimental disseminated candidiasis. The analytical qPCR assay was highly specific and sensitive (10 fg). Cells of C. albicans incubated in Hanks balanced salt solution (+/-10% bovine serum albumin [BSA]) or RPMI (+/-10% BSA) showed a significant release of DNA at T equal to 24 h compared to T equal to 0 h (P < or = 0.01). C. albicans incubated with H-MNCs exhibited a greater release of DNA than C. albicans cells alone over 24 h (P = 0.0001). Rabbits with disseminated candidiasis showed a steady increase of detectable DNA levels in plasma as disease progressed. Plasma cultures showed minimal growth of C. albicans, demonstrating that DNA extracted from plasma reflected fungal cell-free DNA. In summary, these studies of the kinetics of DNA release by C. albicans collectively demonstrate that cell-free fungal DNA is released into the bloodstream of hosts with disseminated candidiasis, that phagocytic cells may play an active role in increasing this release over time, and that plasma is a suitable blood fraction for the detection of C. albicans DNA.
Figures


Similar articles
-
Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis.J Clin Microbiol. 2000 Aug;38(8):3016-21. doi: 10.1128/JCM.38.8.3016-3021.2000. J Clin Microbiol. 2000. PMID: 10921970 Free PMC article.
-
Serum is more suitable than whole blood for diagnosis of systemic candidiasis by nested PCR.J Clin Microbiol. 1999 Apr;37(4):925-30. doi: 10.1128/JCM.37.4.925-930.1999. J Clin Microbiol. 1999. PMID: 10074503 Free PMC article.
-
Detection of Candida albicans in blood by PCR in a rabbit animal model of disseminated candidiasis.Diagn Microbiol Infect Dis. 1999 Jul;34(3):177-83. doi: 10.1016/s0732-8893(99)00036-x. Diagn Microbiol Infect Dis. 1999. PMID: 10403097
-
[Sensitive and specific detection of Candida albicans by polymerase chain reaction].Nihon Rinsho. 1992 Jul;50 Suppl:437-42. Nihon Rinsho. 1992. PMID: 1404936 Review. Japanese. No abstract available.
-
[Advances in the diagnosis of mycoses. New tests detect Candida infections].Lakartidningen. 1998 Feb 18;95(8):738-42. Lakartidningen. 1998. PMID: 9513319 Review. Swedish.
Cited by
-
Gastrointestinal tract distribution of Salmonella enteritidis in orally infected mice with a species-specific fluorescent quantitative polymerase chain reaction.World J Gastroenterol. 2007 Dec 28;13(48):6568-74. doi: 10.3748/wjg.v13.i48.6568. World J Gastroenterol. 2007. PMID: 18161929 Free PMC article.
-
Gastric LTi cells promote lymphoid follicle formation but are limited by IRAK-M and do not alter microbial growth.Mucosal Immunol. 2015 Sep;8(5):1047-59. doi: 10.1038/mi.2014.132. Epub 2015 Jan 21. Mucosal Immunol. 2015. PMID: 25603827 Free PMC article.
-
Candida albicans biofilms and polymicrobial interactions.Crit Rev Microbiol. 2021 Feb;47(1):91-111. doi: 10.1080/1040841X.2020.1843400. Epub 2021 Jan 22. Crit Rev Microbiol. 2021. PMID: 33482069 Free PMC article. Review.
-
Establishment and application of loop-mediated isothermal amplification coupled with nanoparticle-based lateral flow biosensor (LAMP-LFB) for visual and rapid diagnosis of Candida albicans in clinical samples.Front Bioeng Biotechnol. 2022 Nov 7;10:1025083. doi: 10.3389/fbioe.2022.1025083. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36420441 Free PMC article.
-
Establishment and Application of Multiple Cross Displacement Amplification Coupled With Lateral Flow Biosensor (MCDA-LFB) for Visual and Rapid Detection of Candida albicans in Clinical Samples.Front Cell Infect Microbiol. 2019 Apr 16;9:102. doi: 10.3389/fcimb.2019.00102. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31058099 Free PMC article.
References
-
- Benjamin, D. K., Jr., C. Poole, W. J. Steinbach, J. L. Rowen, and T. J. Walsh. 2003. Neonatal candidemia and end-organ damage: a critical appraisal of the literature using meta-analytic techniques. Pediatrics 112:634-640. - PubMed
-
- Bu, R., R. K. Sathiapalan, M. M. Ibrahim, I. Al-Mohsen, E. Almodavar, M. I. Gutierrez, and K. Bhatia. 2005. Monochrome LightCycler PCR assay for detection and quantification of five common species of Candida and Aspergillus. J. Med. Microbiol. 54:243-248. - PubMed
-
- Buchman, T. G., M. Rossier, W. G. Merz, and P. Charache. 1990. Detection of surgical pathogens by in vitro DNA amplification. Part I: rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery 108:338-346. - PubMed
-
- Choi, J. J., C. F. Reich III, and D. S. Pisetsky. 2004. Release of DNA from dead and dying lymphocyte and monocyte cell lines in vitro. Scand. J. Immunol. 60:159-166. - PubMed
-
- Chryssanthou, E., B. Andersson, B. Petrini, S. Lofdahl, and J. Tollemar. 1994. Detection of Candida albicans DNA in serum by polymerase chain reaction. Scand. J. Infect. Dis. 26:479-485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

