Development and validation of a rotor-gene real-time PCR assay for detection, identification, and quantification of Chlamydia trachomatis in a single reaction
- PMID: 16390971
- PMCID: PMC1351959
- DOI: 10.1128/JCM.44.1.206-213.2006
Development and validation of a rotor-gene real-time PCR assay for detection, identification, and quantification of Chlamydia trachomatis in a single reaction
Abstract
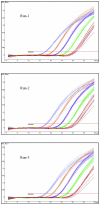
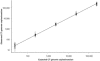
A multitarget real-time PCR (MRT-PCR) for detection of Chlamydia trachomatis DNA was developed and validated. There were three targets for amplification in a single reaction: the cryptic plasmid (CP), the major outer membrane protein (MOMP) gene, and an internal control. The assay had the following characteristics: (i) detection and confirmation of the presence of C. trachomatis DNA in a single reaction, (ii) detection of all genovars of C. trachomatis without any cross-reactivity with pathogenic bacteria or commensal organisms of the oropharynx and genital tract, (iii) a 95% probability of detection with three copies of MOMP and one copy of CP per reaction mixture, (iv) identification of the inhibition of amplification, (v) a quantitative dynamic range of 25 to 250,000 genome copies per reaction mixture, (vi) high intra- and interassay reproducibilities, and (vii) correct identification of all samples in the validation panel. There were 146 COBAS Amplicor PCR (Amplicor PCR)-positive samples and 122 Amplicor PCR-negative samples in the panel. MRT-PCR detected CP DNA alone in 6 (4%) Amplicor PCR-positive samples and both CP and MOMP DNAs in 140 (96%) of 146 Amplicor PCR-positive samples. The quantity of MOMP DNA in 95 (68%) of 140 samples was within the dynamic range of the assay. The median C. trachomatis load in these samples was 321 genome copies per reaction mixture (range, 26 to 40,137 genome copies per reaction mixture). Due to the inclusion of two different C. trachomatis-specific targets, the assay confirmed 259 (97%) of 268 results in a single reaction. This assay could be used in the qualitative format for the routine detection of C. trachomatis and in the quantitative format for study of the pathogenesis of C. trachomatis-associated diseases. The assay demonstrated the potential to eliminate the need for confirmatory testing in almost all samples, thus reducing the turnaround time and the workload.
Figures
References
-
- Aliyu, S. H., R. K. Marriott, M. D. Curran, S. Parmar, N. Bentley, N. M. Brown, J. S. Brazier, and H. Ludlam. 2004. Real-time PCR investigation into the importance of Fusobacterium necrophorum as a cause of acute pharyngitis in general practice. J. Med. Microbiol. 53:1029-1035. - PubMed
-
- An, Q., J. Liu, W. O'Brien, G. Radcliffe, D. Buxton, S. Popoff, W. King, M. Vera-Garcia, L. Lu, J. Shah, J. Klinger, and D. M. Olive. 1995. Comparison of characteristics of Qβ replicase-amplified assay with competitive PCR assay for Chlamydia trachomatis. J. Clin. Microbiol. 33:58-63. - PMC - PubMed
-
- Castriciano, S., K. Lunistra, D. Jang, J. Patel, J. Mahony, J. Kapala, and M. Chernesky. 2002. Accuracy of results obtained by performing a second ligase chain reaction assay and PCR analysis on urine samples with positive or near-cut-off results in the LCx test for Chlamydia trachomatis. J. Clin. Microbiol. 40:2632-2634. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

