The PANE1 gene encodes a novel human minor histocompatibility antigen that is selectively expressed in B-lymphoid cells and B-CLL
- PMID: 16391015
- PMCID: PMC1895781
- DOI: 10.1182/blood-2005-08-3501
The PANE1 gene encodes a novel human minor histocompatibility antigen that is selectively expressed in B-lymphoid cells and B-CLL
Abstract
Minor histocompatibility antigens (mHAg's) are peptides encoded by polymorphic genes that are presented by major histocompatibility complex (MHC) molecules and recognized by T cells in recipients of allogeneic hematopoietic cell transplants. Here we report that an alternative transcript of the proliferation-associated nuclear element 1 (PANE1) gene encodes a novel human leukocyte antigen (HLA)-A(*)0301-restricted mHAg that is selectively expressed in B-lymphoid cells. The antigenic peptide is entirely encoded within a unique exon not present in other PANE1 transcripts. Sequencing of PANE1 alleles in mHAg-positive and mHAg-negative cells demonstrates that differential T-cell recognition is due to a single nucleotide polymorphism within the variant exon that replaces an arginine codon with a translation termination codon. The PANE1 transcript that encodes the mHAg is expressed at high levels in resting CD19(+) B cells and B-lineage chronic lymphocytic leukemia (B-CLL) cells, and at significantly lower levels in activated B cells. Activation of B-CLL cells through CD40 ligand (CD40L) stimulation decreases expression of the mHAg-encoding PANE1 transcript and reciprocally increases expression of PANE1 transcripts lacking the mHAg-encoding exon. These studies suggest distinct roles for different PANE1 isoforms in resting compared with activated CD19(+) cells, and identify PANE1 as a potential therapeutic target in B-CLL.
Figures
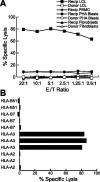

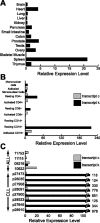
 ) and k (▪) in resting and activated fractions of human peripheral blood leukocytes (B), and in primary ALL and B-CLL samples (C).
) and k (▪) in resting and activated fractions of human peripheral blood leukocytes (B), and in primary ALL and B-CLL samples (C).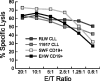

Similar articles
-
Toward targeting B cell cancers with CD4+ CTLs: identification of a CD19-encoded minor histocompatibility antigen using a novel genome-wide analysis.J Exp Med. 2008 Nov 24;205(12):2863-72. doi: 10.1084/jem.20080713. Epub 2008 Nov 10. J Exp Med. 2008. PMID: 19001137 Free PMC article.
-
Identification of a polymorphic gene, BCL2A1, encoding two novel hematopoietic lineage-specific minor histocompatibility antigens.J Exp Med. 2003 Jun 2;197(11):1489-500. doi: 10.1084/jem.20021925. Epub 2003 May 27. J Exp Med. 2003. PMID: 12771180 Free PMC article.
-
Bi-directional allelic recognition of the human minor histocompatibility antigen HB-1 by cytotoxic T lymphocytes.Eur J Immunol. 2002 Oct;32(10):2748-58. doi: 10.1002/1521-4141(2002010)32:10<2748::AID-IMMU2748>3.0.CO;2-T. Eur J Immunol. 2002. PMID: 12355426
-
Secondary anchor polymorphism in the HA-1 minor histocompatibility antigen critically affects MHC stability and TCR recognition.Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3889-94. doi: 10.1073/pnas.0900411106. Epub 2009 Feb 20. Proc Natl Acad Sci U S A. 2009. PMID: 19234124 Free PMC article.
-
Histocompatibility.In: Carreras E, Dufour C, Mohty M, Kröger N, editors. The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies [Internet]. 7th edition. Cham (CH): Springer; 2019. Chapter 9. In: Carreras E, Dufour C, Mohty M, Kröger N, editors. The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies [Internet]. 7th edition. Cham (CH): Springer; 2019. Chapter 9. PMID: 32091748 Free Books & Documents. Review.
Cited by
-
A uniform genomic minor histocompatibility antigen typing methodology and database designed to facilitate clinical applications.PLoS One. 2006 Dec 20;1(1):e42. doi: 10.1371/journal.pone.0000042. PLoS One. 2006. PMID: 17183671 Free PMC article.
-
Adoptive T-cell therapy for B-cell malignancies.Expert Rev Hematol. 2009 Oct;2(5):517-32. doi: 10.1586/ehm.09.47. Expert Rev Hematol. 2009. PMID: 21083018 Free PMC article. Review.
-
Antigen Discovery and Therapeutic Targeting in Hematologic Malignancies.Cancer J. 2017 Mar/Apr;23(2):115-124. doi: 10.1097/PPO.0000000000000257. Cancer J. 2017. PMID: 28410299 Free PMC article. Review.
-
Mechanisms of minor histocompatibility antigen immunogenicity: the role of infinitesimal versus structurally profound polymorphisms.Immunol Res. 2006;36(1-3):33-41. doi: 10.1385/IR:36:1:33. Immunol Res. 2006. PMID: 17337764 Review.
-
High CENPM mRNA expression and its prognostic significance in hepatocellular carcinoma: a study based on data mining.Cancer Cell Int. 2020 Aug 26;20:406. doi: 10.1186/s12935-020-01499-y. eCollection 2020. Cancer Cell Int. 2020. PMID: 32863765 Free PMC article.
References
-
- Chao NJ. Minors come of age: minor histocompatibility antigens and graft-versus-host disease. Biol Blood Marrow Transplant. 2004;10: 215-223. - PubMed
-
- Falkenburg JH, Willemze R. Minor histocompatibility antigens as targets of cellular immunotherapy in leukaemia. Best Pract Res Clin Haematol. 2004;17: 415-425. - PubMed
-
- Bleakley M, Riddell SR. Molecules and mechanisms of the graft-versus-leukaemia effect. Nat Rev Cancer. 2004;4: 371-380. - PubMed
-
- Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307: 1072-1079. - PubMed
-
- de Bueger M, Bakker A, Van Rood JJ, Van der Woude F, Goulmy E. Tissue distribution of human minor histocompatibility antigens: ubiquitous versus restricted tissue distribution indicates heterogeneity among human cytotoxic T lymphocyte-defined non-MHC antigens. J Immunol. 1992; 149: 1788-1794. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

