Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities
- PMID: 16391110
- PMCID: PMC1352197
- DOI: 10.1128/AEM.72.1.702-712.2006
Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities
Abstract
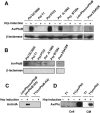
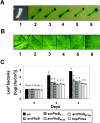
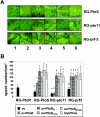
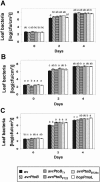
AvrPtoB is a type III effector protein from Pseudomonas syringae pv. tomato that physically interacts with the tomato Pto kinase and, depending on the host genotype, either elicits or suppresses programmed cell death associated with plant immunity. We reported previously that avrPtoB-related sequences are present in diverse gram-negative phytopathogenic bacteria. Here we describe characterization of avrPtoB homologs from P. syringae pv. tomato T1, PT23, and JL1065, P. syringae pv. syringae B728a, and P. syringae pv. maculicola ES4326. The avrPtoB homolog from P. syringae pv. maculicola, hopPmaL, was identified previously. The four new genes identified in this study are designated avrPtoB(T1), avrPtoB(PT23), avrPtoB(JL1065), and avrPtoB(B728a). The AvrPtoB homologs exhibit 52 to 66% amino acid identity with AvrPtoB. Transcripts of each of the avrPtoB homologs were detected in the Pseudomonas strains from which they were isolated. Proteins encoded by the homologs were detected in all strains except P. syringae pv. tomato T1, suggesting that T1 suppresses accumulation of AvrPtoB(T1). All of the homologs interacted with the Pto kinase in a yeast two-hybrid system and elicited a Pto-dependent defense response when they were delivered into leaf cells by DC3000DeltaavrPtoDeltaavrPtoB, a P. syringae pv. tomato strain with a deletion of both avrPto and avrPtoB. Like AvrPtoB, all of the homologs enhanced the ability of DC3000DeltaavrPtoDeltaavrPtoB to form lesions on leaves of two susceptible tomato lines. With the exception of HopPmaL which lacks the C-terminal domain, all AvrPtoB homologs suppressed programmed cell death elicited by the AvrPto-Pto interaction in an Agrobacterium-mediated transient assay. Thus, despite their divergent sequences, AvrPtoB homologs from diverse P. syringae pathovars have conserved avirulence and virulence activities similar to AvrPtoB activity.
Figures


Similar articles
-
Pto- and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse pseudomonas syringae pathovars to infect tomato.Mol Plant Microbe Interact. 2007 Jul;20(7):806-15. doi: 10.1094/MPMI-20-7-0806. Mol Plant Microbe Interact. 2007. PMID: 17601168
-
The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants.Plant J. 2007 Nov;52(4):595-614. doi: 10.1111/j.1365-313X.2007.03259.x. Epub 2007 Aug 31. Plant J. 2007. PMID: 17764515 Free PMC article.
-
An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato.Mol Plant Microbe Interact. 2005 Jan;18(1):43-51. doi: 10.1094/MPMI-18-0043. Mol Plant Microbe Interact. 2005. PMID: 15672817
-
AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity.FEMS Microbiol Lett. 2005 Apr 1;245(1):1-8. doi: 10.1016/j.femsle.2005.02.025. FEMS Microbiol Lett. 2005. PMID: 15796972 Review.
-
Effector-triggered immunity mediated by the Pto kinase.Trends Plant Sci. 2011 Mar;16(3):132-40. doi: 10.1016/j.tplants.2010.11.001. Epub 2010 Nov 26. Trends Plant Sci. 2011. PMID: 21112235 Review.
Cited by
-
Pseudomonas syringae pv. syringae uses proteasome inhibitor syringolin A to colonize from wound infection sites.PLoS Pathog. 2013 Mar;9(3):e1003281. doi: 10.1371/journal.ppat.1003281. Epub 2013 Mar 28. PLoS Pathog. 2013. PMID: 23555272 Free PMC article.
-
Pseudomonas syringae pv. actinidiae Type III Effectors Localized at Multiple Cellular Compartments Activate or Suppress Innate Immune Responses in Nicotiana benthamiana.Front Plant Sci. 2017 Dec 20;8:2157. doi: 10.3389/fpls.2017.02157. eCollection 2017. Front Plant Sci. 2017. PMID: 29326748 Free PMC article.
-
A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity.Nature. 2007 Jul 19;448(7151):370-4. doi: 10.1038/nature05966. Nature. 2007. PMID: 17637671 Free PMC article.
-
Nonhost resistance of tomato to the bean pathogen Pseudomonas syringae pv. syringae B728a is due to a defective E3 ubiquitin ligase domain in avrptobb728a.Mol Plant Microbe Interact. 2013 Apr;26(4):387-97. doi: 10.1094/MPMI-08-12-0190-R. Mol Plant Microbe Interact. 2013. PMID: 23252461 Free PMC article.
-
Race-specific genotypes of Pseudomonas syringae pv. tomato are defined by the presence of mobile DNA elements within the genome.Front Plant Sci. 2023 Jul 5;14:1197706. doi: 10.3389/fpls.2023.1197706. eCollection 2023. Front Plant Sci. 2023. PMID: 37476164 Free PMC article.
References
-
- Abramovitch, R. B., and G. B. Martin. 2005. AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol. Lett. 245:1-8. - PubMed
-
- Abramovitch, R. B., and G. B. Martin. 2004. Strategies used by bacterial pathogens to suppress plant defense. Curr. Opin. Plant Biol. 7:356-364. - PubMed
-
- Badel, J. L., K. Nomura, S. Bandyopadhyay, R. Shimizu, A. Collmer, and S. Y. He. 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49:1239-1251. - PubMed
-
- Bretz, J., L. Losada, K. Lisboa, and S. W. Hutcheson. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45:397-409. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

