Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method
- PMID: 16391112
- PMCID: PMC1352191
- DOI: 10.1128/AEM.72.1.723-732.2006
Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method
Abstract
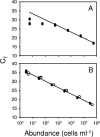
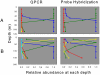
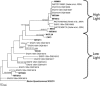
The cyanobacterium Prochlorococcus numerically dominates the photosynthetic community in the tropical and subtropical regions of the world's oceans. Six evolutionary lineages of Prochlorococcus have been described, and their distinctive physiologies and genomes indicate that these lineages are "ecotypes" and should have different oceanic distributions. Two methods recently developed to quantify these ecotypes in the field, probe hybridization and quantitative PCR (QPCR), have shown that this is indeed the case. To facilitate a global investigation of these ecotypes, we modified our QPCR protocol to significantly increase its speed, sensitivity, and accessibility and validated the method in the western and eastern North Atlantic Ocean. We showed that all six ecotypes had distinct distributions that varied with depth and location, and, with the exception of the deeper waters at the western North Atlantic site, the total Prochlorococcus counts determined by QPCR matched the total counts measured by flow cytometry. Clone library analyses of the deeper western North Atlantic waters revealed ecotypes that are not represented in the culture collections with which the QPCR primers were designed, explaining this discrepancy. Finally, similar patterns of relative ecotype abundance were obtained in QPCR and probe hybridization analyses of the same field samples, which could allow comparisons between studies.
Figures


References
-
- Ahlgren, N. A., G. Rocap, and S. W. Chisholm. Measurement of Prochlorococcus ecotypes using real-time PCR reveals different abundances of genotypes with similar light physiologies. Environ. Microbiol., in press. - PubMed
-
- Campbell, L., H. Liu, H. A. Nolla, and D. Vaulot. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991-1994 ENSO event. Deep-Sea Res. Part I 44:167-192.
-
- Cavender-Bares, K. K., S. L. Frankel, and S. W. Chisholm. 1998. A dual sheath flow cytometer for shipboard analyses of phytoplankton communities from the oligotrophic oceans. Limnol. Oceanogr. 43:1383-1388.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

