Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides
- PMID: 16391113
- PMCID: PMC1352245
- DOI: 10.1128/AEM.72.1.733-744.2006
Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides
Abstract
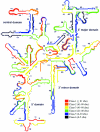
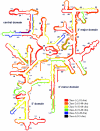
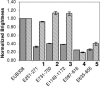
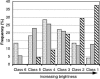
rRNA accessibility is a major sensitivity issue limiting the design of working probes for fluorescence in situ hybridization (FISH). Previous studies empirically highlighted the accessibility of target sites on rRNA maps by grouping probes into six classes according to their brightness levels. In this study, a recently proposed mechanistic model of FISH, based on the thermodynamics of secondary nucleic acid interactions, was used to evaluate the accessibility of the 16S rRNA of Escherichia coli to fluorescein-labeled oligonucleotides when thermodynamic and kinetic barriers were eliminated. To cover the entire 16S rRNA, 109 probes were designed with an average thermodynamic affinity (DeltaGo (overall)) of -13.5 kcal/mol. Fluorescence intensity was measured by flow cytometry, and a brightness threshold between classes 3 and 4 was used as the requirement for proof of accessibility. While 46% of the probes were above this threshold with conventional 3-h hybridizations, extending the incubation period to 96 h dramatically increased the fraction of bright probes to 86%. Insufficient thermodynamic affinity and/or fluorophore quenching was demonstrated to cause the low fluorescence intensity of the remaining 14% of the probes. In the end, it was proven that every nucleotide in the 16S rRNA of E. coli could be targeted with a bright probe and, therefore, that there were no truly inaccessible target regions in the 16S rRNA. Based on our findings and mechanistic modeling, a rational design strategy involving DeltaGo(overall), hybridization kinetics, and fluorophore quenching is recommended for the development of bright probes.
Figures


References
-
- Amann, R. 1996. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, D. J. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
-
- Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. - PubMed
-
- Behrens, S., B. M. Fuchs, and R. Amann. 2004. The effect of nucleobase-specific fluorescence quenching on in situ hybridization with rRNA-targeted oligonucleotide probes. Syst. Appl. Microbiol. 27:565-572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

