Modulation of rat cecal microbiota by administration of raffinose and encapsulated Bifidobacterium breve
- PMID: 16391119
- PMCID: PMC1352276
- DOI: 10.1128/AEM.72.1.784-792.2006
Modulation of rat cecal microbiota by administration of raffinose and encapsulated Bifidobacterium breve
Abstract
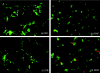
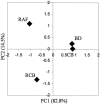
To investigate the effects of administration of raffinose and encapsulated Bifidobacterium breve JCM 1192T cells on the rat cecal microbiota, in a preclinical synbiotic study groups of male WKAH/Hkm Slc rats were fed for 3 weeks with four different test diets: basal diet (group BD), basal diet supplemented with raffinose (group RAF), basal diet supplemented with encapsulated B. breve (group CB), and basal diet supplemented with both raffinose and encapsulated B. breve (group RCB). The bacterial populations in cecal samples were determined by fluorescence in situ hybridization (FISH) and terminal restriction fragment length polymorphism (T-RFLP). B. breve cells were detected only in the RCB group and accounted for about 6.3% of the total cells as determined by FISH analysis. B. breve was also detected only in the RCB group by T-RFLP analysis. This was in contrast to the CB group, in which no B. breve signals were detected by either FISH or T-RFLP. Increases in the sizes of the populations of Bifidobacterium animalis, a Bifidobacterium indigenous to the rat, were observed in the RAF and RCB groups. Principal-component analysis of T-RFLP results revealed significant alterations in the bacterial populations of rats in the RAF and RCB groups; the population in the CB group was similar to that in the control group (group BD). To the best of our knowledge, these results provide the first clear picture of the changes in the rat cecal microbiota in response to synbiotic administration.
Figures


Similar articles
-
Population dynamics of Bifidobacterium species in human feces during raffinose administration monitored by fluorescence in situ hybridization-flow cytometry.Appl Environ Microbiol. 2006 Dec;72(12):7739-47. doi: 10.1128/AEM.01777-06. Epub 2006 Oct 20. Appl Environ Microbiol. 2006. PMID: 17056700 Free PMC article.
-
Synbiotic promotion of epithelial proliferation by orally ingested encapsulated Bifidobacterium breve and raffinose in the small intestine of rats.Mol Nutr Food Res. 2009 May;53 Suppl 1:S62-7. doi: 10.1002/mnfr.200800041. Mol Nutr Food Res. 2009. PMID: 18837471
-
Diets supplemented with chickpea or its main oligosaccharide component raffinose modify faecal microbial composition in healthy adults.Benef Microbes. 2010 Jun;1(2):197-207. doi: 10.3920/BM2009.0027. Benef Microbes. 2010. PMID: 21831757 Clinical Trial.
-
Different utilization of glucose and raffinose in Bifidobacterium breve and Bifidobacterium animalis.Folia Microbiol (Praha). 2006;51(4):320-4. doi: 10.1007/BF02931824. Folia Microbiol (Praha). 2006. PMID: 17007436
-
Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species.FEMS Microbiol Lett. 2005 Feb 15;243(2):417-23. doi: 10.1016/j.femsle.2005.01.002. FEMS Microbiol Lett. 2005. PMID: 15686844
Cited by
-
Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation.Appl Environ Microbiol. 2012 Jul;78(14):4984-94. doi: 10.1128/AEM.00588-12. Epub 2012 May 11. Appl Environ Microbiol. 2012. PMID: 22582061 Free PMC article.
-
Population dynamics of Bifidobacterium species in human feces during raffinose administration monitored by fluorescence in situ hybridization-flow cytometry.Appl Environ Microbiol. 2006 Dec;72(12):7739-47. doi: 10.1128/AEM.01777-06. Epub 2006 Oct 20. Appl Environ Microbiol. 2006. PMID: 17056700 Free PMC article.
-
Effects of feeding potato pulp on cholesterol metabolism and its association with cecal conditions in rats.Plant Foods Hum Nutr. 2011 Nov;66(4):401-7. doi: 10.1007/s11130-011-0255-z. Plant Foods Hum Nutr. 2011. PMID: 21948633
-
Bifidobacterium breve as a delivery vector of IL-24 gene therapy for head and neck squamous cell carcinoma in vivo.Gene Ther. 2017 Nov;24(11):699-705. doi: 10.1038/gt.2017.74. Epub 2017 Aug 14. Gene Ther. 2017. PMID: 28805796
-
Characterization of housing-related spontaneous variations of gut microbiota and expression of toll-like receptors 2 and 4 in rats.Microb Ecol. 2010 Oct;60(3):691-702. doi: 10.1007/s00248-010-9737-z. Epub 2010 Aug 18. Microb Ecol. 2010. PMID: 20717659
References
-
- Alander, M., J. Mättö, W. Kneifel, M. Johansson, B. Kögler, R. Crittenden, T. Mattila-Sandholm, and M. Saarela. 2001. Effect of galacto-oligosaccharide supplementation on human faecal microflora and on survival and persistence of Bifidobacterium lactis Bb-12 in the gastrointestinal tract. Int. Dairy J. 11:817-825.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

