Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi
- PMID: 16391138
- PMCID: PMC1352221
- DOI: 10.1128/AEM.72.1.932-936.2006
Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi
Abstract
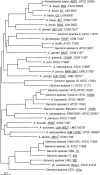
Genotypic and phenotypic analyses were carried out to clarify the taxonomic position of the naturally transformable Acinetobacter sp. strain ADP1. Transfer tDNA-PCR fingerprinting, 16S rRNA gene sequence analysis, and selective restriction fragment amplification (amplified fragment length polymorphism analysis) indicate that strain ADP1 and a second transformable strain, designated 93A2, are members of the newly described species Acinetobacter baylyi. Transformation assays demonstrate that the A. baylyi type strain B2(T) and two other originally identified members of the species (C5 and A7) also have the ability to undergo natural transformation at high frequencies, confirming that these five strains belong to a separate species of the genus Acinetobacter, characterized by the high transformability of its strains that have been cultured thus far.
Figures


Similar articles
-
Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemolytic organisms isolated from humans.Int J Syst Evol Microbiol. 2009 Jan;59(Pt 1):118-24. doi: 10.1099/ijs.0.001230-0. Int J Syst Evol Microbiol. 2009. PMID: 19126734
-
Description of Acinetobacter venetianus ex Di Cello et al. 1997 sp. nov.Int J Syst Evol Microbiol. 2009 Jun;59(Pt 6):1376-81. doi: 10.1099/ijs.0.003541-0. Int J Syst Evol Microbiol. 2009. PMID: 19502319
-
Unique organization of the 16S-23S intergenic spacer regions of strains of Acinetobacter baylyi provides a means for its identification from other Acinetobacter species.J Microbiol Methods. 2008 Jun;73(3):227-36. doi: 10.1016/j.mimet.2008.03.003. Epub 2008 Mar 14. J Microbiol Methods. 2008. PMID: 18436316
-
Acinetobacter baylyi ADP1: transforming the choice of model organism.IUBMB Life. 2011 Dec;63(12):1075-80. doi: 10.1002/iub.530. Epub 2011 Oct 27. IUBMB Life. 2011. PMID: 22034222 Review.
-
Acinetobacter baylyi ADP1-naturally competent for synthetic biology.Essays Biochem. 2021 Jul 26;65(2):309-318. doi: 10.1042/EBC20200136. Essays Biochem. 2021. PMID: 33769448 Review.
Cited by
-
Analysis of storage lipid accumulation in Alcanivorax borkumensis: Evidence for alternative triacylglycerol biosynthesis routes in bacteria.J Bacteriol. 2007 Feb;189(3):918-28. doi: 10.1128/JB.01292-06. Epub 2006 Nov 22. J Bacteriol. 2007. PMID: 17122340 Free PMC article.
-
Diversity within the O-linked protein glycosylation systems of acinetobacter species.Mol Cell Proteomics. 2014 Sep;13(9):2354-70. doi: 10.1074/mcp.M114.038315. Epub 2014 Jun 10. Mol Cell Proteomics. 2014. PMID: 24917611 Free PMC article.
-
Engineering Acinetobacter baylyi ADP1 for mevalonate production from lignin-derived aromatic compounds.Metab Eng Commun. 2021 May 24;13:e00173. doi: 10.1016/j.mec.2021.e00173. eCollection 2021 Dec. Metab Eng Commun. 2021. PMID: 34430203 Free PMC article.
-
The genome of Polaromonas sp. strain JS666: insights into the evolution of a hydrocarbon- and xenobiotic-degrading bacterium, and features of relevance to biotechnology.Appl Environ Microbiol. 2008 Oct;74(20):6405-16. doi: 10.1128/AEM.00197-08. Epub 2008 Aug 22. Appl Environ Microbiol. 2008. PMID: 18723656 Free PMC article.
-
CatM regulation of the benABCDE operon: functional divergence of two LysR-type paralogs in Acinetobacter baylyi ADP1.Appl Environ Microbiol. 2006 Mar;72(3):1749-58. doi: 10.1128/AEM.72.3.1749-1758.2006. Appl Environ Microbiol. 2006. PMID: 16517618 Free PMC article.
References
-
- Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. - PMC - PubMed
-
- Bouvet, P. J., and P. A. Grimont. 1987. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur Microbiol. 138:569-578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

