Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes
- PMID: 16391231
- PMCID: PMC1356099
- DOI: 10.1101/gad.1381306
Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes
Abstract
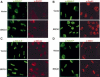
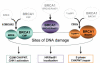
The BRCA1 gene product and its stoichiometric binding partner, BARD1, play a vital role in the cellular response to DNA damage. However, how they acquire specific biochemical functions after DNA damage is poorly understood. Following exposure to genotoxic stress, DNA damage-specific interactions were observed between BRCA1/BARD1 and the DNA damage-response proteins, TopBP1 and Mre11/Rad50/NBS1. Two distinct DNA damage-dependent super complexes emerged; their activation was dependent, in part, on the actions of specific checkpoint kinases, and each super complex contributed to a distinctive aspect of the DNA damage response. The results support a new, multifactorial model that describes how genotoxic stress enables BRCA1 to execute a diverse set of DNA damage-response functions.
Figures





References
-
- Aladjem M.I., Rodewald, L.W., Kolman, J.L., and Wahl, G.M. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281: 1005–1009. - PubMed
-
- Avni D., Yang, H., Martelli, F., Hofmann, F., ElShamy, W.M., Ganesan, S., Scully, R., and Livingston, D.M. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12: 735–746. - PubMed
-
- Bakkenist C.J. and Kastan, M.B. 2004. Initiating cellular stress responses. Cell 118: 9–17. - PubMed
-
- Bartek J. and Lukas, J. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429. - PubMed
-
- Bartek J., Lukas, C., and Lukas, J. 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell. Biol. 5: 792–804. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
