Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli
- PMID: 16391237
- PMCID: PMC1356105
- DOI: 10.1101/gad.366606
Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli
Abstract
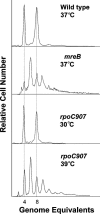
The actin-like MreB cytoskeletal protein and RNA polymerase (RNAP) have both been suggested to provide the force for chromosome segregation. Here, we identify MreB and RNAP as in vivo interaction partners. The interaction was confirmed using in vitro purified components. We also present convincing evidence that MreB and RNAP are both required for chromosome segregation in Escherichia coli. MreB is required for origin and bulk DNA segregation, whereas RNAP is required for bulk DNA, terminus, and possibly also for origin segregation. Furthermore, flow cytometric analyses show that MreB depletion and inactivation of RNAP confer virtually identical and highly unusual chromosome segregation defects. Thus, our results raise the possibility that the MreB-RNAP interaction is functionally important for chromosome segregation.
Figures




References
-
- Bellomy G.R. and Record Jr., M.T. 1990. Stable DNA loops in vivo and in vitro: Roles in gene regulation at a distance and in biophysical characterization of DNA. Prog. Nucleic Acid Res. Mol. Biol. 39: 81–128. - PubMed
-
- Ben-Yehuda S., Rudner, D.Z., and Losick, R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536. - PubMed
-
- Ben-Yehuda S., Fujita, M., Liu, X.S., Gorbatyuk, B., Skoko, D., Yan, J., Marko, J.F., Liu, J.S., Eichenberger, P., Rudner, D.Z., et al. 2005. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17: 773–782. - PubMed
-
- Bettinger B.T., Gilbert, D.M., and Amberg, D.C. 2004. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 5: 410–415. - PubMed
-
- Blagoev B., Kratchmarova, I., Ong, S.E., Nielsen, M., Foster, L.J., and Mann, M. 2003. A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nat. Biotechnol. 21: 315–318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
