Alternative splicing regulation at tandem 3' splice sites
- PMID: 16394021
- PMCID: PMC1325015
- DOI: 10.1093/nar/gkj408
Alternative splicing regulation at tandem 3' splice sites
Abstract
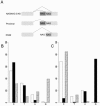
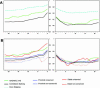
Alternative splicing (AS) constitutes a major mechanism creating protein diversity in humans. Previous bioinformatics studies based on expressed sequence tag and mRNA data have identified many AS events that are conserved between humans and mice. Of these events, approximately 25% are related to alternative choices of 3' and 5' splice sites. Surprisingly, half of all these events involve 3' splice sites that are exactly 3 nt apart. These tandem 3' splice sites result from the presence of the NAGNAG motif at the acceptor splice site, recently reported to be widely spread in the human genome. Although the NAGNAG motif is common in human genes, only a small subset of sites with this motif is confirmed to be involved in AS. We examined the NAGNAG motifs and observed specific features such as high sequence conservation of the motif, high conservation of approximately 30 bp at the intronic regions flanking the 3' splice site and overabundance of cis-regulatory elements, which are characteristic of alternatively spliced tandem acceptor sites and can distinguish them from the constitutive sites in which the proximal NAG splice site is selected. Our findings imply that AS at tandem splice sites and constitutive splicing of the distal NAG are highly regulated.
Figures
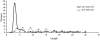

References
-
- Mercatante D., Kole R. Modification of alternative splicing pathways as a potential approach to chemotherapy. Pharmacol. Ther. 2000;85:237–243. - PubMed
-
- Sugnet C.W., Kent W.J., Ares M., Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004:66–77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

