The V260I mutation in fission yeast alpha-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint
- PMID: 16394105
- PMCID: PMC1382329
- DOI: 10.1091/mbc.e05-08-0802
The V260I mutation in fission yeast alpha-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint
Abstract
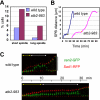
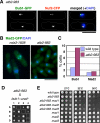
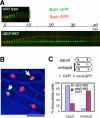
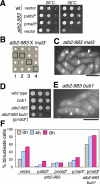
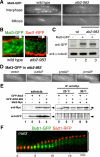
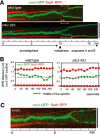
We have identified a novel temperature-sensitive mutant of fission yeast alpha-tubulin Atb2 (atb2-983) that contains a single amino acid substitution (V260I). Atb2-983 is incorporated into the microtubules, and their overall structures are not altered noticeably, but microtubule dynamics is compromised during interphase. atb2-983 displays a high rate of chromosome missegregation and is synthetically lethal with deletions in a subset of spindle checkpoint genes including bub1, bub3, and mph1, but not with mad1, mad2, and mad3. During early mitosis in this mutant, Bub1, but not Mad2, remains for a prolonged period in the kinetochores that are situated in proximity to one of the two SPBs (spindle pole bodies). High dosage mal3(+), encoding EB1 homologue, rescues atb2-983, suggesting that Mal3 function is compromised. Consistently, Mal3 localization and binding between Mal3 and Atb2-983 are impaired significantly, and a mal3 single mutant, such as atb2-983, displays prolonged Bub1 kinetochore localization. Furthermore in atb2-983 back-and-forth centromere oscillation during prometaphase is abolished. Intriguingly, this oscillation still occurs in the mal3 mutant, indicating that there is another defect independent of Mal3. These results show that microtubule dynamics is important for coordinated execution of mitotic events, in which Mal3 plays a vital role.
Figures



Similar articles
-
Mal3, the fission yeast EB1 homologue, cooperates with Bub1 spindle checkpoint to prevent monopolar attachment.EMBO Rep. 2005 Dec;6(12):1194-200. doi: 10.1038/sj.embor.7400540. EMBO Rep. 2005. PMID: 16179942 Free PMC article.
-
Novel mad2 alleles isolated in a Schizosaccharomyces pombe gamma-tubulin mutant are defective in metaphase arrest activity, but remain functional for chromosome stability in unperturbed mitosis.Genetics. 2007 Apr;175(4):1571-84. doi: 10.1534/genetics.106.061309. Epub 2007 Feb 4. Genetics. 2007. PMID: 17277378 Free PMC article.
-
Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1.Proc Natl Acad Sci U S A. 2012 Jan 3;109(1):209-14. doi: 10.1073/pnas.1114647109. Epub 2011 Dec 19. Proc Natl Acad Sci U S A. 2012. PMID: 22184248 Free PMC article.
-
Cooperation of EB1-Mal3 and the Bub1 spindle checkpoint.Cell Cycle. 2006 Jan;5(1):27-30. doi: 10.4161/cc.5.1.2262. Epub 2006 Jan 18. Cell Cycle. 2006. PMID: 16294010 Review.
-
Spatiotemporal control of spindle disassembly in fission yeast.Cell Mol Life Sci. 2019 Sep;76(18):3543-3551. doi: 10.1007/s00018-019-03139-9. Epub 2019 May 25. Cell Mol Life Sci. 2019. PMID: 31129857 Free PMC article. Review.
Cited by
-
Microtubules in Microorganisms: How Tubulin Isotypes Contribute to Diverse Cytoskeletal Functions.Front Cell Dev Biol. 2022 Jul 5;10:913809. doi: 10.3389/fcell.2022.913809. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35865635 Free PMC article. Review.
-
MAARS: a novel high-content acquisition software for the analysis of mitotic defects in fission yeast.Mol Biol Cell. 2017 Jun 15;28(12):1601-1611. doi: 10.1091/mbc.E16-10-0723. Epub 2017 Apr 27. Mol Biol Cell. 2017. PMID: 28450455 Free PMC article.
-
Mutations in alpha-tubulin confer dinitroaniline resistance at a cost to microtubule function.Mol Biol Cell. 2007 Dec;18(12):4711-20. doi: 10.1091/mbc.e07-04-0379. Epub 2007 Sep 19. Mol Biol Cell. 2007. PMID: 17881728 Free PMC article.
-
The dual role of fission yeast Tbc1/cofactor C orchestrates microtubule homeostasis in tubulin folding and acts as a GAP for GTPase Alp41/Arl2.Mol Biol Cell. 2013 Jun;24(11):1713-24, S1-8. doi: 10.1091/mbc.E12-11-0792. Epub 2013 Apr 10. Mol Biol Cell. 2013. PMID: 23576550 Free PMC article.
-
Motor neuron synapse and axon defects in a C. elegans alpha-tubulin mutant.PLoS One. 2010 Mar 11;5(3):e9655. doi: 10.1371/journal.pone.0009655. PLoS One. 2010. PMID: 20300184 Free PMC article.
References
-
- Asakawa, K., and Toda, T. (2006). Cooperation of EB1-Mal3 and the Bub1 spindle checkpoint. Cell Cycle 5, 27–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

