Ddx42p--a human DEAD box protein with RNA chaperone activities
- PMID: 16397294
- PMCID: PMC1325199
- DOI: 10.1093/nar/gkj403
Ddx42p--a human DEAD box protein with RNA chaperone activities
Abstract
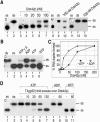
The human gene ddx42 encodes a human DEAD box protein highly homologous to the p68 subfamily of RNA helicases. In HeLa cells, two ddx42 poly(A)+ RNA species were detected both encoding the nuclear localized 938 amino acid Ddx42p polypeptide. Ddx42p has been heterologously expressed and its biochemical properties characterized. It is an RNA binding protein, and ATP and ADP modulate its RNA binding affinity. Ddx42p is an NTPase with a preference for ATP, the hydrolysis of which is enhanced by various RNA substrates. It acts as a non-processive RNA helicase. Interestingly, RNA unwinding by Ddx42p is promoted in the presence of a single-strand (ss) binding protein (T4gp32). Ddx42p, particularly in the ADP-bound form (the state after ATP hydrolysis), also mediates efficient annealing of complementary RNA strands thereby displacing the ss binding protein. Ddx42p therefore represents the first example of a human DEAD box protein possessing RNA helicase, protein displacement and RNA annealing activities. The adenosine nucleotide cofactor bound to Ddx42p apparently acts as a switch that controls the two opposing activities: ATP triggers RNA strand separation, whereas ADP triggers annealing of complementary RNA strands.
Figures

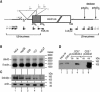
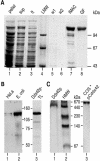
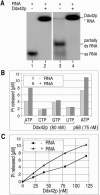
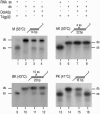
References
-
- Shiratori A., Shibata T., Arisawa M., Hanaoka F., Marakami Y., Eki T. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and northern analysis. Yeast. 1999;15:219–253. - PubMed
-
- Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. - PubMed
-
- Hickson I.D. RecQ helicases: caretakers of the genome. Nature Rev. Cancer. 2003;3:169–178. - PubMed
-
- Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim. Biophys. Acta. 2004;1704:37–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

