RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP
- PMID: 16397502
- PMCID: PMC1560093
- DOI: 10.1038/nature04331
RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP
Abstract
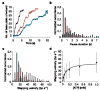
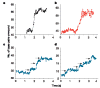
Helicases are a ubiquitous class of enzymes involved in nearly all aspects of DNA and RNA metabolism. Despite recent progress in understanding their mechanism of action, limited resolution has left inaccessible the detailed mechanisms by which these enzymes couple the rearrangement of nucleic acid structures to the binding and hydrolysis of ATP. Observing individual mechanistic cycles of these motor proteins is central to understanding their cellular functions. Here we follow in real time, at a resolution of two base pairs and 20 ms, the RNA translocation and unwinding cycles of a hepatitis C virus helicase (NS3) monomer. NS3 is a representative superfamily-2 helicase essential for viral replication, and therefore a potentially important drug target. We show that the cyclic movement of NS3 is coordinated by ATP in discrete steps of 11 +/- 3 base pairs, and that actual unwinding occurs in rapid smaller substeps of 3.6 +/- 1.3 base pairs, also triggered by ATP binding, indicating that NS3 might move like an inchworm. This ATP-coupling mechanism is likely to be applicable to other non-hexameric helicases involved in many essential cellular functions. The assay developed here should be useful in investigating a broad range of nucleic acid translocation motors.
Figures


References
-
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. - PubMed
-
- Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:751–771. - PubMed
-
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

