Developmentally regulated DNA methylation in Dictyostelium discoideum
- PMID: 16400165
- PMCID: PMC1360260
- DOI: 10.1128/EC.5.1.18-25.2006
Developmentally regulated DNA methylation in Dictyostelium discoideum
Abstract
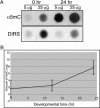
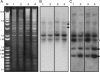
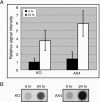
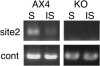
Methylation of cytosine residues in DNA plays a critical role in the silencing of gene expression, organization of chromatin structure, and cellular differentiation of eukaryotes. Previous studies failed to detect 5-methylcytosine in Dictyostelium genomic DNA, but the recent sequencing of the Dictyostelium genome revealed a candidate DNA methyltransferase gene (dnmA). The genome sequence also uncovered an unusual distribution of potential methylation sites, CpG islands, throughout the genome. DnmA belongs to the Dnmt2 subfamily and contains all the catalytic motifs necessary for cytosine methyltransferases. Dnmt2 activity is typically weak in Drosophila melanogaster, mouse, and human cells and the gene function in these systems is unknown. We have investigated the methylation status of Dictyostelium genomic DNA with antibodies raised against 5-methylcytosine and detected low levels of the modified nucleotide. We also found that DNA methylation increased during development. We searched the genome for potential methylation sites and found them in retrotransposable elements and in several other genes. Using Southern blot analysis with methylation-sensitive and -insensitive restriction endonucleases, we found that the DIRS retrotransposon and the guaB gene were indeed methylated. We then mutated the dnmA gene and found that DNA methylation was reduced to about 50% of the wild-type level. The mutant cells exhibited morphological defects in late development, indicating that DNA methylation has a regulatory role in Dictyostelium development. Our findings establish a role for a Dnmt2 methyltransferase in eukaryotic development.
Figures


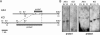

References
-
- Adachi, H., T. Hasebe, K. Yoshinaga, T. Ohta, and K. Sutoh. 1994. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205:1808-1814. - PubMed
-
- Adams, R. L. 1996. DNA methylation, p. 33-66. In E. E. Bitter (ed.), Principles of medical biology, vol. 5. JAI Press, Inc., New York, N.Y.
-
- Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. - PubMed
-
- Bestor, T. H. 1998. The host defence function of genomic methylation patterns. Novartis Found. Symp. 214:187-199, 228-232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

