Cell cycle-dependent localization and properties of a second mitochondrial DNA ligase in Crithidia fasciculata
- PMID: 16400168
- PMCID: PMC1360255
- DOI: 10.1128/EC.5.1.54-61.2006
Cell cycle-dependent localization and properties of a second mitochondrial DNA ligase in Crithidia fasciculata
Abstract
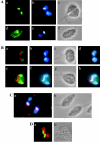
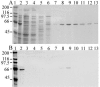
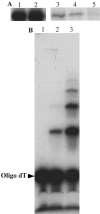
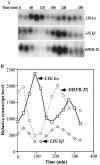
The mitochondrial DNA in kinetoplastid protozoa is contained in a single highly condensed structure consisting of thousands of minicircles and approximately 25 maxicircles. The disk-shaped structure is termed kinetoplast DNA (kDNA) and is located in the mitochondrial matrix near the basal body. We have previously identified a mitochondrial DNA ligase (LIG kbeta) in the trypanosomatid Crithidia fasciculata that localizes to antipodal sites flanking the kDNA disk where several other replication proteins are localized. We describe here a second mitochondrial DNA ligase (LIG kalpha). LIG kalpha localizes to the kinetoplast primarily in cells that have completed mitosis and contain either a dividing kinetoplast or two newly divided kinetoplasts. Essentially all dividing or newly divided kinetoplasts show localization of LIG kalpha. The ligase is present on both faces of the kDNA disk and at a high level in the kinetoflagellar zone of the mitochondrial matrix. Cells containing a single nucleus show localization of the LIG kalpha to the kDNA but at a much lower frequency. The mRNA level of LIG kalpha varies during the cell cycle out of phase with that of LIG kbeta. LIG kalpha transcript levels are maximal during the phase when cells contain two nuclei, whereas LIG kbeta transcript levels are maximal during S phase. The LIG kalpha protein decays with a half-life of 100 min in the absence of protein synthesis. The periodic expression of the LIG kalpha transcript and the instability of the LIG kalpha protein suggest a possible role of the ligase in regulating minicircle replication.
Figures

Similar articles
-
Mitochondrial DNA ligases of Trypanosoma brucei.Eukaryot Cell. 2005 Apr;4(4):765-74. doi: 10.1128/EC.4.4.765-774.2005. Eukaryot Cell. 2005. PMID: 15821136 Free PMC article.
-
Mitochondrial DNA ligase in Crithidia fasciculata.Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4361-6. doi: 10.1073/pnas.0305705101. Epub 2004 Mar 2. Proc Natl Acad Sci U S A. 2004. PMID: 15070723 Free PMC article.
-
Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata.Mol Cell Biol. 1996 Feb;16(2):564-76. doi: 10.1128/MCB.16.2.564. Mol Cell Biol. 1996. PMID: 8552084 Free PMC article.
-
The structure and replication of kinetoplast DNA.Curr Mol Med. 2004 Sep;4(6):623-47. doi: 10.2174/1566524043360096. Curr Mol Med. 2004. PMID: 15357213 Review.
-
Fellowship of the rings: the replication of kinetoplast DNA.Trends Parasitol. 2005 Aug;21(8):363-9. doi: 10.1016/j.pt.2005.06.008. Trends Parasitol. 2005. PMID: 15967722 Review.
Cited by
-
Identification of new kinetoplast DNA replication proteins in trypanosomatids based on predicted S-phase expression and mitochondrial targeting.Eukaryot Cell. 2007 Dec;6(12):2303-10. doi: 10.1128/EC.00284-07. Epub 2007 Oct 26. Eukaryot Cell. 2007. PMID: 17965251 Free PMC article.
-
Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids.Trends Parasitol. 2011 Jan;27(1):23-30. doi: 10.1016/j.pt.2010.06.011. Epub 2010 Jul 6. Trends Parasitol. 2011. PMID: 20609625 Free PMC article. Review.
-
TbPIF8, a Trypanosoma brucei protein related to the yeast Pif1 helicase, is essential for cell viability and mitochondrial genome maintenance.Mol Microbiol. 2012 Feb;83(3):471-85. doi: 10.1111/j.1365-2958.2011.07938.x. Epub 2012 Jan 4. Mol Microbiol. 2012. PMID: 22220754 Free PMC article.
-
Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes.Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19250-5. doi: 10.1073/pnas.0706858104. Epub 2007 Nov 28. Proc Natl Acad Sci U S A. 2007. PMID: 18048338 Free PMC article.
-
A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei.Eukaryot Cell. 2011 Mar;10(3):445-54. doi: 10.1128/EC.00308-10. Epub 2011 Jan 21. Eukaryot Cell. 2011. PMID: 21257796 Free PMC article.
References
-
- Abu-Elneel, K., I. Kapeller, and J. Shlomai. 1999. Universal minicircle sequence-binding protein, a sequence-specific DNA-binding protein that recognizes the two replication origins of the kinetoplast DNA minicircle. J. Biol. Chem. 274:13419-13426. - PubMed
-
- Birkenmeyer, L., and D. S. Ray. 1986. Replication of kinetoplast DNA in isolated kinetoplasts from Crithidia fasciculata. J. Biol. Chem. 261:2362-2368. - PubMed
-
- Birkenmeyer, L., H. Sugisaki, and D. S. Ray. 1987. Structural characterization of site-specific discontinuities associated with replication origins of minicircle DNA from Crithidia fasciculata. J. Biol. Chem. 262:2384-2392. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

