Pair of unusual GCN5 histone acetyltransferases and ADA2 homologues in the protozoan parasite Toxoplasma gondii
- PMID: 16400169
- PMCID: PMC1360262
- DOI: 10.1128/EC.5.1.62-76.2006
Pair of unusual GCN5 histone acetyltransferases and ADA2 homologues in the protozoan parasite Toxoplasma gondii
Abstract
GCN5 is a histone acetyltransferase (HAT) essential for development in mammals and critical to stress responses in yeast. The protozoan parasite Toxoplasma gondii is a serious opportunistic pathogen. The study of epigenetics and gene expression in this ancient eukaryote has pharmacological relevance and may facilitate the understanding of these processes in higher eukaryotes. Here we show that the disruption of T. gondii GCN5 yields viable parasites, which were subsequently employed in a proteomics study to identify gene products affected by its loss. Promoter analysis of these TgGCN5-dependent genes, which were mostly parasite specific, reveals a conserved T-rich element. The loss of TgGCN5 does not attenuate virulence in an in vivo mouse model. We also discovered that T. gondii is the only invertebrate reported to date possessing a second GCN5 (TgGCN5-B). TgGCN5-B harbors a strikingly divergent N-terminal domain required for nuclear localization. Despite high homology between the HAT domains, the two TgGCN5s exhibit differing substrate specificities. In contrast to TgGCN5-A, which exclusively targets lysine 18 of H3, TgGCN5-B acetylates multiple lysines in the H3 tail. We also identify two ADA2 homologues that interact differently with the TgGCN5s. TgGCN5-B has the potential to compensate for TgGCN5-A, which probably arose from a gene duplication unique to T. gondii. Our work reveals an unexpected complexity in the GCN5 machinery of this primitive eukaryote.
Figures
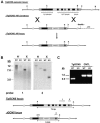
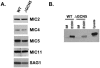







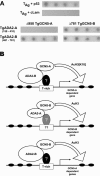
Similar articles
-
Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii.J Biol Chem. 2005 Feb 18;280(7):5902-8. doi: 10.1074/jbc.M410656200. Epub 2004 Dec 9. J Biol Chem. 2005. PMID: 15591057
-
Regions of intrinsic disorder help identify a novel nuclear localization signal in Toxoplasma gondii histone acetyltransferase TgGCN5-B.Mol Biochem Parasitol. 2011 Feb;175(2):192-5. doi: 10.1016/j.molbiopara.2010.10.009. Epub 2010 Nov 3. Mol Biochem Parasitol. 2011. PMID: 21055425 Free PMC article.
-
Cloning and analysis of a Toxoplasma gondii histone acetyltransferase: a novel chromatin remodelling factor in Apicomplexan parasites.Nucleic Acids Res. 1999 Nov 15;27(22):4344-52. doi: 10.1093/nar/27.22.4344. Nucleic Acids Res. 1999. PMID: 10536141 Free PMC article.
-
The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194629. doi: 10.1016/j.bbagrm.2020.194629. Epub 2020 Sep 2. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32890768 Free PMC article. Review.
-
Toxoplasma histone acetylation remodelers as novel drug targets.Expert Rev Anti Infect Ther. 2012 Oct;10(10):1189-201. doi: 10.1586/eri.12.100. Expert Rev Anti Infect Ther. 2012. PMID: 23199404 Free PMC article. Review.
Cited by
-
Previously Unidentified Histone H1-Like Protein Is Involved in Cell Division and Ribosome Biosynthesis in Toxoplasma gondii.mSphere. 2022 Dec 21;7(6):e0040322. doi: 10.1128/msphere.00403-22. Epub 2022 Dec 5. mSphere. 2022. PMID: 36468865 Free PMC article.
-
O-fucosylated glycoproteins form assemblies in close proximity to the nuclear pore complexes of Toxoplasma gondii.Proc Natl Acad Sci U S A. 2016 Oct 11;113(41):11567-11572. doi: 10.1073/pnas.1613653113. Epub 2016 Sep 23. Proc Natl Acad Sci U S A. 2016. PMID: 27663739 Free PMC article.
-
Involvement of a Toxoplasma gondii chromatin remodeling complex ortholog in developmental regulation.PLoS One. 2011;6(5):e19570. doi: 10.1371/journal.pone.0019570. Epub 2011 May 31. PLoS One. 2011. PMID: 21655329 Free PMC article.
-
Modifications at K31 on the lateral surface of histone H4 contribute to genome structure and expression in apicomplexan parasites.Elife. 2017 Nov 4;6:e29391. doi: 10.7554/eLife.29391. Elife. 2017. PMID: 29101771 Free PMC article.
-
Toxoplasma: the next 100years.Microbes Infect. 2008 Jul;10(9):978-84. doi: 10.1016/j.micinf.2008.07.015. Epub 2008 Jul 10. Microbes Infect. 2008. PMID: 18672085 Free PMC article. Review.
References
-
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. - PubMed
-
- Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. - PubMed
-
- Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. - PubMed
-
- Bhatti, M. M., and W. J. Sullivan, Jr. 2005. Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J. Biol. Chem. 280:5902-5908. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

