Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1
- PMID: 16400172
- PMCID: PMC1360268
- DOI: 10.1128/EC.5.1.103-111.2006
Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1
Abstract
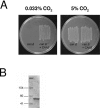
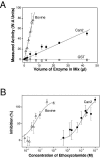
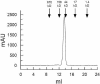
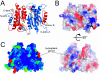
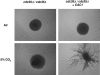
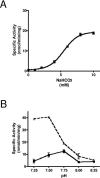
Cryptococcus neoformans, a fungal pathogen of humans, causes fatal meningitis in immunocompromised patients. Its virulence is mainly determined by the elaboration of a polysaccharide capsule surrounding its cell wall. During its life, C. neoformans is confronted with and responds to dramatic variations in CO2 concentrations; one important morphological change triggered by the shift from its natural habitat (0.033% CO2) to infected hosts (5% CO2) is the induction of capsule biosynthesis. In cells, CO2 is hydrated to bicarbonate in a spontaneous reaction that is accelerated by carbonic anhydrases. Here we show that C. neoformans contains two beta-class carbonic anhydrases, Can1 and Can2. We further demonstrate that CAN2, but not CAN1, is abundantly expressed and essential for the growth of C. neoformans in its natural environment, where CO2 concentrations are limiting. Structural studies reveal that Can2 forms a homodimer in solution. Our data reveal Can2 to be the main carbonic anhydrase and suggest a physiological role for bicarbonate during C. neoformans growth. Bicarbonate directly activates the C. neoformans Cac1 adenylyl cyclase required for capsule synthesis. We show that this specific activation is optimal at physiological pH.
Figures




Similar articles
-
Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans.J Mol Biol. 2009 Jan 30;385(4):1207-20. doi: 10.1016/j.jmb.2008.11.037. Epub 2008 Nov 27. J Mol Biol. 2009. PMID: 19071134
-
Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence.Curr Biol. 2005 Nov 22;15(22):2013-20. doi: 10.1016/j.cub.2005.09.047. Curr Biol. 2005. PMID: 16303560
-
Comparative transcriptome analysis of the CO2 sensing pathway via differential expression of carbonic anhydrase in Cryptococcus neoformans.Genetics. 2010 Aug;185(4):1207-19. doi: 10.1534/genetics.110.118315. Epub 2010 Jun 1. Genetics. 2010. PMID: 20516494 Free PMC article.
-
CO2 sensing in fungi and beyond.Curr Opin Microbiol. 2006 Dec;9(6):572-8. doi: 10.1016/j.mib.2006.09.003. Epub 2006 Oct 11. Curr Opin Microbiol. 2006. PMID: 17045514 Review.
-
A matter of structure: structural comparison of fungal carbonic anhydrases.Appl Microbiol Biotechnol. 2014 Oct;98(20):8433-41. doi: 10.1007/s00253-014-5993-z. Epub 2014 Aug 12. Appl Microbiol Biotechnol. 2014. PMID: 25109265 Review.
Cited by
-
Network analysis exposes core functions in major lifestyles of fungal and oomycete plant pathogens.BMC Genomics. 2019 Dec 26;20(1):1020. doi: 10.1186/s12864-019-6409-3. BMC Genomics. 2019. PMID: 31878885 Free PMC article.
-
Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans.J Clin Invest. 2006 Jun;116(6):1651-9. doi: 10.1172/JCI27890. J Clin Invest. 2006. PMID: 16741577 Free PMC article.
-
Fungal adaptation to the mammalian host: it is a new world, after all.Curr Opin Microbiol. 2008 Dec;11(6):511-6. doi: 10.1016/j.mib.2008.09.018. Epub 2008 Nov 3. Curr Opin Microbiol. 2008. PMID: 18955154 Free PMC article. Review.
-
Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans.Cell Microbiol. 2012 Apr;14(4):500-16. doi: 10.1111/j.1462-5822.2011.01735.x. Epub 2012 Jan 9. Cell Microbiol. 2012. PMID: 22151739 Free PMC article.
-
Cryptococcal virulence: beyond the usual suspects.J Clin Invest. 2006 Jun;116(6):1481-3. doi: 10.1172/JCI28842. J Clin Invest. 2006. PMID: 16741574 Free PMC article.
References
-
- Abadi, J., S. Nachman, A. B. Kressel, and L. Pirofski. 1999. Cryptococcosis in children with AIDS. Clin. Infect. Dis. 28:309-313. - PubMed
-
- Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75-84. - PMC - PubMed
-
- Amoroso, G., L. Morell-Avrahov, D. Muller, K. Klug, and D. Sultemeyer. 2005. The gene NCE103 (YNL036w) from Saccharomyces cerevisiae encodes a functional carbonic anhydrase and its transcription is regulated by the concentration of inorganic carbon in the medium. Mol. Microbiol. 56:549-558. - PubMed
-
- Anonymous. Cryptococcus neoformans Serotype A Database. [Online.] http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans/.
-
- Bahn, Y. S., G. M. Cox, J. R. Perfect, and J. Heitman. 2005. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 15:2013-2020. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

