Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils
- PMID: 16401079
- PMCID: PMC1435828
- DOI: 10.1021/bi051952q
Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils
Abstract
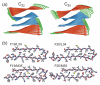
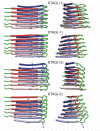
We describe solid-state nuclear magnetic resonance (NMR) measurements on fibrils formed by the 40-residue beta-amyloid peptide associated with Alzheimer's disease (Abeta(1-40)) that place constraints on the identity and symmetry of contacts between in-register, parallel beta-sheets in the fibrils. We refer to these contacts as internal and external quaternary contacts, depending on whether they are within a single molecular layer or between molecular layers. The data include (1) two-dimensional 13C-13C NMR spectra that indicate internal quaternary contacts between side chains of L17 and F19 and side chains of I32, L34, and V36, as well as external quaternary contacts between side chains of I31 and G37; (2) two-dimensional 15N-13C NMR spectra that indicate external quaternary contacts between the side chain of M35 and the peptide backbone at G33; (3) measurements of magnetic dipole-dipole couplings between the side chain carboxylate group of D23 and the side chain amine group of K28 that indicate salt bridge interactions. Isotopic dilution experiments allow us to make distinctions between intramolecular and intermolecular contacts. On the basis of these data and previously determined structural constraints from solid-state NMR and electron microscopy, we construct full molecular models using restrained molecular dynamics simulations and restrained energy minimization. These models apply to Abeta(1-40) fibrils grown with gentle agitation. We also present evidence for different internal quaternary contacts in Abeta(1-40) fibrils grown without agitation, which are morphologically distinct.
Figures






References
-
- Sunde M, Blake CCF. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q. Rev. Biophys. 1998;31:1–39. - PubMed
-
- Tycko R. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struct. Biol. 2004;14:96–103. - PubMed
-
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. - PubMed
-
- Wickner RB, Edskes HK, Ross ED, Pierce MM, Baxa U, Brachmann A, Shewmaker F. Prion genetics: New rules for a new kind of gene. Annu. Rev. Genet. 2004;38:681–707. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

