Sulfur K-edge XAS and DFT calculations on nitrile hydratase: geometric and electronic structure of the non-heme iron active site
- PMID: 16402841
- PMCID: PMC4485618
- DOI: 10.1021/ja0549695
Sulfur K-edge XAS and DFT calculations on nitrile hydratase: geometric and electronic structure of the non-heme iron active site
Abstract
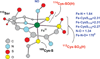
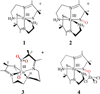
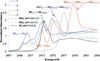
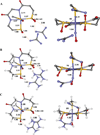
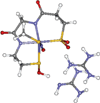
The geometric and electronic structure of the active site of the non-heme iron enzyme nitrile hydratase (NHase) is studied using sulfur K-edge XAS and DFT calculations. Using thiolate (RS(-))-, sulfenate (RSO(-))-, and sulfinate (RSO(2)(-))-ligated model complexes to provide benchmark spectral parameters, the results show that the S K-edge XAS is sensitive to the oxidation state of S-containing ligands and that the spectrum of the RSO(-) species changes upon protonation as the S-O bond is elongated (by approximately 0.1 A). These signature features are used to identify the three cysteine residues coordinated to the low-spin Fe(III) in the active site of NHase as CysS(-), CysSOH, and CysSO(2)(-) both in the NO-bound inactive form and in the photolyzed active form. These results are correlated to geometry-optimized DFT calculations. The pre-edge region of the X-ray absorption spectrum is sensitive to the Z(eff) of the Fe and reveals that the Fe in [FeNO](6) NHase species has a Z(eff) very similar to that of its photolyzed Fe(III) counterpart. DFT calculations reveal that this results from the strong pi back-bonding into the pi antibonding orbital of NO, which shifts significant charge from the formally t(2)(6) low-spin metal to the coordinated NO.
Figures





References
-
- Endo I, Nojiri M, Tsujimura M, Nakasako M, Nagashima S, Yohda M, Odaka M. J. Inorg. Biochem. 2001;83:247–253. - PubMed
-
- Yamada H, Kobayashi M. Biosci. Biotechnol. Biochem. 1996;60:1391–1400. - PubMed
-
- Nagasawa T, Takeuchi K, Yamada H. Eur. J. Biochem. 1991;196:581–589. - PubMed
-
- Payne MS, Wu S, Fallon RD, Tudor G, Stieglitz B, Turner IM, Nelson MJ. Biochemistry. 1997;36:5447–5454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

