Ca2+-ATPases in non-failing and failing heart: evidence for a novel cardiac sarco/endoplasmic reticulum Ca2+-ATPase 2 isoform (SERCA2c)
- PMID: 16402920
- PMCID: PMC1422767
- DOI: 10.1042/BJ20051427
Ca2+-ATPases in non-failing and failing heart: evidence for a novel cardiac sarco/endoplasmic reticulum Ca2+-ATPase 2 isoform (SERCA2c)
Abstract
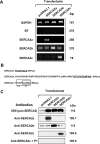
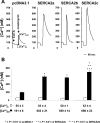
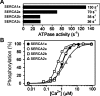
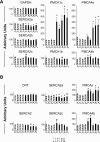
We recently documented the expression of a novel human mRNA variant encoding a yet uncharacterized SERCA [SR (sarcoplasmic reticulum)/ER (endoplasmic reticulum) Ca2+-ATPase] protein, SERCA2c [Gélébart, Martin, Enouf and Papp (2003) Biochem. Biophys. Res. Commun. 303, 676-684]. In the present study, we have analysed the expression and functional characteristics of SERCA2c relative to SERCA2a and SERCA2b isoforms upon their stable heterologous expression in HEK-293 cells (human embryonic kidney 293 cells). All SERCA2 proteins induced an increased Ca2+ content in the ER of intact transfected cells. In microsomes prepared from transfected cells, SERCA2c showed a lower apparent affinity for cytosolic Ca2+ than SERCA2a and a catalytic turnover rate similar to SERCA2b. We further demonstrated the expression of the endogenous SERCA2c protein in protein lysates isolated from heart left ventricles using a newly generated SERCA2c-specific antibody. Relative to the known uniform distribution of SERCA2a and SERCA2b in cardiomyocytes of the left ventricle tissue, SERCA2c was only detected in a confined area of cardiomyocytes, in close proximity to the sarcolemma. This finding led us to explore the expression of the presently known cardiac Ca2+-ATPase isoforms in heart failure. Comparative expression of SERCAs and PMCAs (plasma-membrane Ca2+-ATPases) was performed in four nonfailing hearts and five failing hearts displaying mixed cardiomyopathy and idiopathic dilated cardiomyopathies. Relative to normal subjects, cardiomyopathic patients express more PMCAs than SERCA2 proteins. Interestingly, SERCA2c expression was significantly increased (166+/-26%) in one patient. Taken together, these results demonstrate the expression of the novel SERCA2c isoform in the heart and may point to a still unrecognized role of PMCAs in cardiomyopathies.
Figures



Similar articles
-
Multiple and diverse coexpression, location, and regulation of additional SERCA2 and SERCA3 isoforms in nonfailing and failing human heart.J Mol Cell Cardiol. 2010 Apr;48(4):633-44. doi: 10.1016/j.yjmcc.2009.11.012. Epub 2009 Dec 4. J Mol Cell Cardiol. 2010. PMID: 19962989 Review.
-
Compartmentalized expression of three novel sarco/endoplasmic reticulum Ca2+ATPase 3 isoforms including the switch to ER stress, SERCA3f, in non-failing and failing human heart.Cell Calcium. 2009 Feb;45(2):144-54. doi: 10.1016/j.ceca.2008.08.002. Epub 2008 Oct 22. Cell Calcium. 2009. PMID: 18947868
-
Three novel sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) 3 isoforms. Expression, regulation, and function of the membranes of the SERCA3 family.J Biol Chem. 2002 Jul 5;277(27):24442-52. doi: 10.1074/jbc.M202011200. Epub 2002 Apr 15. J Biol Chem. 2002. PMID: 11956212
-
Sarcoplasmic reticulum Ca2+ATPase and phospholamban mRNA and protein levels in end-stage heart failure due to ischemic or dilated cardiomyopathy.J Mol Med (Berl). 1996 Jun;74(6):321-32. doi: 10.1007/BF00207509. J Mol Med (Berl). 1996. PMID: 8862513
-
Gene knockout studies of Ca2+-transporting ATPases.Eur J Biochem. 2000 Sep;267(17):5284-90. doi: 10.1046/j.1432-1327.2000.01568.x. Eur J Biochem. 2000. PMID: 10951186 Review.
Cited by
-
Benefit of SERCA2a gene transfer to vascular endothelial and smooth muscle cells: a new aspect in therapy of cardiovascular diseases.Curr Vasc Pharmacol. 2013 Jul;11(4):465-79. doi: 10.2174/1570161111311040010. Curr Vasc Pharmacol. 2013. PMID: 23905641 Free PMC article. Review.
-
Comparison of transcriptomic landscapes of different lamb muscles using RNA-Seq.PLoS One. 2018 Jul 24;13(7):e0200732. doi: 10.1371/journal.pone.0200732. eCollection 2018. PLoS One. 2018. PMID: 30040835 Free PMC article.
-
Advances in gene-based therapy for heart failure.J Cardiovasc Transl Res. 2008 Jun;1(2):127-36. doi: 10.1007/s12265-008-9022-4. Epub 2008 May 29. J Cardiovasc Transl Res. 2008. PMID: 20559907 Review.
-
Structure-function relationships and modifications of cardiac sarcoplasmic reticulum Ca2+-transport.Physiol Res. 2021 Dec 30;70(Suppl4):S443-S470. doi: 10.33549/physiolres.934805. Physiol Res. 2021. PMID: 35199536 Free PMC article. Review.
-
Apoptosis in differentiating C2C12 muscle cells selectively targets Bcl-2-deficient myotubes.Apoptosis. 2014 Jan;19(1):42-57. doi: 10.1007/s10495-013-0922-7. Apoptosis. 2014. PMID: 24129924 Free PMC article.
References
-
- Berridge M. J. The versatility and complexity of calcium signalling. Novartis Found. Symp. 2001;239:52–64. - PubMed
-
- Wuytack F., Raeymaekers L., Missiaen L. Molecular physiology of the SERCA and SPCA pumps. Cell Calcium. 2002;32:279–305. - PubMed
-
- Shull G. E., Okunade G., Liu L. H., Kozel P., Periasamy M., Lorenz J. N., Prasad V. Physiological functions of plasma membrane and intracellular Ca2+ pumps revealed by analysis of null mutants. Ann. N.Y. Acad. Sci. 2003;986:453–460. - PubMed
-
- Strehler E. E., Treiman M. Calcium pumps of plasma membrane and cell interior. Curr. Mol. Med. 2004;4:323–335. - PubMed
-
- Bobe R., Bredoux R., Corvazier E., Lacabaratz-Porret C., Martin V., Kovacs T., Enouf J. How many Ca2+ATPase isoforms in a cell type?. A growing membrane protein family exemplified in platelets. Platelets. 2005;16:133–150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

