Combined megaplex TCR isolation and SMART-based real-time quantitation methods for quantitating antigen-specific T cell clones in mycobacterial infection
- PMID: 16403511
- PMCID: PMC2884368
- DOI: 10.1016/j.jim.2005.09.009
Combined megaplex TCR isolation and SMART-based real-time quantitation methods for quantitating antigen-specific T cell clones in mycobacterial infection
Abstract
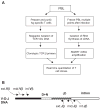
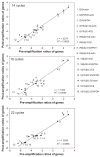
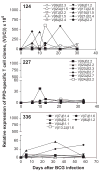
Despite recent advances in measuring cellular immune responses, the quantitation of antigen-specific T cell clones in infections or diseases remains challenging. Here, we employed combined megaplex TCR isolation and SMART-based real-time quantitation methods to quantitate numerous antigen-specific T cell clones using limited amounts of specimens. The megaplex TCR isolation covered the repertoire comprised of recombinants from 24 Vbeta families and 13 Jbeta segments, and allowed us to isolate TCR VDJ clonotypic sequences from one or many PPD-specific IFNgamma-producing T cells that were purified by flow cytometry sorting. The SMART amplification technique was then validated for its capacity to proportionally enrich cellular TCR mRNA/cDNA for real-time quantitation of large numbers of T cell clones. SMART amplified cDNA was shown to maintain relative expression levels of TCR genes when compared to unamplified cDNA. While the SMART-based real-time quantitative PCR conferred a detection limit of 10(-5) to 10(-6) antigen-specific T cells, the clonotypic primers specifically amplified and quantitated the target clone TCR but discriminated other clones that differed by >or=2 bases in the DJ regions. Furthermore, the combined megaplex TCR isolation and SMART-based real-time quantiation methods allowed us to quantitate large numbers of PPD-specific IFNgamma-producing T cell clones using as few as 2 x 10(6) PBMC collected weekly after mycobacterial infection. This assay system may be useful for studies of antigen-specific T cell clones in tumors, autoimmune and infectious diseases.
Figures
Similar articles
-
TCR repertoire, clonal dominance, and pulmonary trafficking of mycobacterium-specific CD4+ and CD8+ T effector cells in immunity against tuberculosis.J Immunol. 2010 Oct 1;185(7):3940-7. doi: 10.4049/jimmunol.1001222. Epub 2010 Aug 30. J Immunol. 2010. PMID: 20805423 Free PMC article.
-
Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire.J Exp Med. 1994 Apr 1;179(4):1261-71. doi: 10.1084/jem.179.4.1261. J Exp Med. 1994. PMID: 8145043 Free PMC article.
-
Multiple sclerosis: T-cell receptor expression in distinct brain regions.Brain. 2007 Nov;130(Pt 11):2789-99. doi: 10.1093/brain/awm214. Epub 2007 Sep 21. Brain. 2007. PMID: 17890278
-
Application of the molecular analysis of the T-cell receptor repertoire in the study of immune-mediated hematologic diseases.Hematology. 2003 Jun;8(3):173-81. doi: 10.1080/1024533031000107505. Hematology. 2003. PMID: 12745651 Review.
-
Immune-mediated bone marrow failure syndromes of progenitor and stem cells: molecular analysis of cytotoxic T cell clones.Folia Histochem Cytobiol. 2007;45(1):5-14. Folia Histochem Cytobiol. 2007. PMID: 17378239 Review.
Cited by
-
The past, present, and future of immune repertoire biology - the rise of next-generation repertoire analysis.Front Immunol. 2013 Nov 27;4:413. doi: 10.3389/fimmu.2013.00413. Front Immunol. 2013. PMID: 24348479 Free PMC article. Review.
-
The clonal composition of human CD4+CD25+Foxp3+ cells determined by a comprehensive DNA-based multiplex PCR for TCRB gene rearrangements.J Immunol Methods. 2007 Apr 10;321(1-2):107-20. doi: 10.1016/j.jim.2007.01.005. Epub 2007 Feb 2. J Immunol Methods. 2007. PMID: 17316678 Free PMC article.
-
Clonal immune responses of Mycobacterium-specific γδ T cells in tuberculous and non-tuberculous tissues during M. tuberculosis infection.PLoS One. 2012;7(2):e30631. doi: 10.1371/journal.pone.0030631. Epub 2012 Feb 1. PLoS One. 2012. PMID: 22319574 Free PMC article.
-
Immune gene networks of mycobacterial vaccine-elicited cellular responses and immunity.J Infect Dis. 2007 Jan 1;195(1):55-69. doi: 10.1086/509895. Epub 2006 Nov 22. J Infect Dis. 2007. PMID: 17152009 Free PMC article.
-
Missing: a diagnostic technique to enumerate antigen-specific T cells.Crit Rev Oncol Hematol. 2012 Aug;83(2):276-82. doi: 10.1016/j.critrevonc.2011.11.002. Epub 2011 Dec 3. Crit Rev Oncol Hematol. 2012. PMID: 22137827 Free PMC article. Review.
References
-
- Akatsuka Y, Martin EG, Madonik A, Barsoukov AA, Hansen JA. Rapid screening of T-cell receptor (TCR) variable gene usage by multiplex PCR: application for assessment of clonal composition. Tissue Antigens. 1999;53:122. - PubMed
-
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94. - PubMed
-
- Chen ZW, Kou ZC, Lekutis C, Shen L, Zhou D, Halloran M, Li J, Sodroski J, Lee-Parritz D, Letvin NL. T cell receptor V beta repertoire in an acute infection of rhesus monkeys with simian immunodeficiency viruses and a chimeric simian–human immunodeficiency virus. J Exp Med. 1995;182:21. - PMC - PubMed
-
- Chen ZW, Shen L, Regan JD, Kou Z, Ghim SH, Letvin NL. The T cell receptor gene usage by simian immunodeficiency virus gag-specific cytotoxic T lymphocytes in rhesus monkeys. J Immunol. 1996;156:1469. - PubMed
-
- Chen ZW, Li Y, Zeng X, Kuroda MJ, Schmitz JE, Shen Y, Lai X, Shen L, Letvin NL. The TCR repertoire of an immunodominant CD8+ T lymphocyte population. J Immunol. 2001;166:4525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

