Mechanism of the Class I KDPG aldolase
- PMID: 16403639
- PMCID: PMC3315828
- DOI: 10.1016/j.bmc.2005.12.022
Mechanism of the Class I KDPG aldolase
Abstract
In vivo, 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase catalyzes the reversible, stereospecific retro-aldol cleavage of KDPG to pyruvate and D-glyceraldehyde-3-phosphate. The enzyme is a lysine-dependent (Class I) aldolase that functions through the intermediacy of a Schiff base. Here, we propose a mechanism for this enzyme based on crystallographic studies of wild-type and mutant aldolases. The three dimensional structure of KDPG aldolase from the thermophile Thermotoga maritima was determined to 1.9A. The structure is the standard alpha/beta barrel observed for all Class I aldolases. At the active site Lys we observe clear density for a pyruvate Schiff base. Density for a sulfate ion bound in a conserved cluster of residues close to the Schiff base is also observed. We have also determined the structure of a mutant of Escherichia coli KDPG aldolase in which the proposed general acid/base catalyst has been removed (E45N). One subunit of the trimer contains density suggesting a trapped pyruvate carbinolamine intermediate. All three subunits contain a phosphate ion bound in a location effectively identical to that of the sulfate ion bound in the T. maritima enzyme. The sulfate and phosphate ions experimentally locate the putative phosphate binding site of the aldolase and, together with the position of the bound pyruvate, facilitate construction of a model for the full-length KDPG substrate complex. The model requires only minimal positional adjustments of the experimentally determined covalent intermediate and bound anion to accommodate full-length substrate. The model identifies the key catalytic residues of the protein and suggests important roles for two observable water molecules. The first water molecule remains bound to the enzyme during the entire catalytic cycle, shuttling protons between the catalytic glutamate and the substrate. The second water molecule arises from dehydration of the carbinolamine and serves as the nucleophilic water during hydrolysis of the enzyme-product Schiff base. The second water molecule may also mediate the base-catalyzed enolization required to form the carbon nucleophile, again bridging to the catalytic glutamate. Many aspects of this mechanism are observed in other Class I aldolases and suggest a mechanistically and, perhaps, evolutionarily related family of aldolases distinct from the N-acetylneuraminate lyase (NAL) family.
Figures
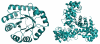


Similar articles
-
Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution.Proc Natl Acad Sci U S A. 2001 Mar 27;98(7):3679-84. doi: 10.1073/pnas.071380898. Proc Natl Acad Sci U S A. 2001. PMID: 11274385 Free PMC article.
-
Characterization and crystal structure of Escherichia coli KDPGal aldolase.Bioorg Med Chem. 2008 Jan 15;16(2):710-20. doi: 10.1016/j.bmc.2007.10.043. Epub 2007 Oct 18. Bioorg Med Chem. 2008. PMID: 17981470 Free PMC article.
-
Structures of Zymomonas 2-Keto-3-Deoxy-6-Phosphogluconate Aldolase with and without a Substrate Analog at the Phosphate-Binding Loop.J Microbiol Biotechnol. 2018 Aug 28;28(8):1339-1345. doi: 10.4014/jmb.1804.04009. J Microbiol Biotechnol. 2018. PMID: 29943554
-
Sweet siblings with different faces: the mechanisms of FBP and F6P aldolase, transaldolase, transketolase and phosphoketolase revisited in light of recent structural data.Bioorg Chem. 2014 Dec;57:263-280. doi: 10.1016/j.bioorg.2014.09.001. Epub 2014 Sep 16. Bioorg Chem. 2014. PMID: 25267444 Review.
-
Current state of and need for enzyme engineering of 2-deoxy-D-ribose 5-phosphate aldolases and its impact.Appl Microbiol Biotechnol. 2021 Aug;105(16-17):6215-6228. doi: 10.1007/s00253-021-11462-0. Epub 2021 Aug 19. Appl Microbiol Biotechnol. 2021. PMID: 34410440 Free PMC article. Review.
Cited by
-
Improving upon nature: active site remodeling produces highly efficient aldolase activity toward hydrophobic electrophilic substrates.Biochemistry. 2012 Feb 28;51(8):1658-68. doi: 10.1021/bi201899b. Epub 2012 Feb 16. Biochemistry. 2012. PMID: 22316217 Free PMC article.
-
Identification of 2-oxohistidine Interacting Proteins Using E. coli Proteome Chips.Mol Cell Proteomics. 2016 Dec;15(12):3581-3593. doi: 10.1074/mcp.M116.060806. Epub 2016 Sep 19. Mol Cell Proteomics. 2016. PMID: 27644758 Free PMC article.
-
Design of multi-scale protein complexes by hierarchical building block fusion.Nat Commun. 2021 Apr 16;12(1):2294. doi: 10.1038/s41467-021-22276-z. Nat Commun. 2021. PMID: 33863889 Free PMC article.
-
Complete and cooperative in vitro assembly of computationally designed self-assembling protein nanomaterials.Nat Commun. 2021 Feb 9;12(1):883. doi: 10.1038/s41467-021-21251-y. Nat Commun. 2021. PMID: 33563988 Free PMC article.
-
Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Streptococcus suis serotype 2.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Nov 1;64(Pt 11):997-9. doi: 10.1107/S1744309108028340. Epub 2008 Oct 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18997324 Free PMC article.
References
-
- Takayama S, McGarvey GJ, Wong CH. Annu. Rev. Microbiol. 1997;51:285. - PubMed
-
- Zannetti MT, Walter C, Knorst M, Fessner WD. Chem.-Eur. J. 1999;5:1882.
-
- Heine A, Luz JG, Wong CH, Wilson IA. J. Mol. Biol. 2004;343:1019. - PubMed
-
- Racker E. J. Biol. Chem. 1952;196:347. - PubMed
-
- Liu JQ, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H. J. Mol. Catal. B—Enzym. 2000;10:107.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

