HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells
- PMID: 16407133
- PMCID: PMC1334651
- DOI: 10.1073/pnas.0507631103
HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells
Erratum in
- Proc Natl Acad Sci U S A. 2006 Jun 6;103(23):8906
Abstract
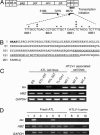
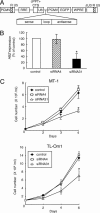
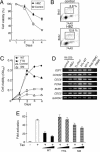
Human T cell leukemia virus type I (HTLV-I) causes adult T cell leukemia (ATL) in 2-5% of carriers after a long latent period. An HTLV-I encoded protein, Tax, induces proliferation and inhibits apoptosis, resulting in clonal proliferation of infected cells. However, tax gene expression in ATL cells is disrupted by several mechanisms, including genetic changes in the tax gene and DNA methylation/deletion of the 5' long terminal repeat (LTR). Because Tax is the major target of cytotoxic T-lymphocytes in vivo, loss of Tax expression should enable ATL cells to escape the host immune system. The 5' LTR of HTLV-I is frequently hypermethylated or deleted in ATL cells, whereas the 3' LTR remains unmethylated and intact, suggesting the involvement of the 3' LTR in leukemogenesis. Here we show that a gene encoded by the minus strand of the HTLV-I proviral genome, HTLV-I basic leucine zipper factor (HBZ), is transcribed from 3'-LTR in all ATL cells. Suppression of HBZ gene transcription by short interfering RNA inhibits proliferation of ATL cells. In addition, HBZ gene expression promotes proliferation of a human T cell line. Analyses of T cell lines transfected with mutated HBZ genes showed that HBZ promotes T cell proliferation in its RNA form, whereas HBZ protein suppresses Tax-mediated viral transcription through the 5' LTR. Thus, the single HBZ gene has bimodal functions in two different molecular forms. The growth-promoting activity of HBZ RNA likely plays an important role in oncogenesis by HTLV-I.
Figures


Similar articles
-
[Adult T-cell leukemia induced by HTLV-1: before and after HBZ].Med Sci (Paris). 2010 Apr;26(4):391-6. doi: 10.1051/medsci/2010264391. Med Sci (Paris). 2010. PMID: 20412744 Review. French.
-
Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells.Retrovirology. 2008 Apr 22;5:34. doi: 10.1186/1742-4690-5-34. Retrovirology. 2008. PMID: 18426605 Free PMC article.
-
HTLV-1 bZIP factor dysregulates the Wnt pathways to support proliferation and migration of adult T-cell leukemia cells.Oncogene. 2013 Sep 5;32(36):4222-30. doi: 10.1038/onc.2012.450. Epub 2012 Oct 8. Oncogene. 2013. PMID: 23045287
-
ATF3, an HTLV-1 bZip factor binding protein, promotes proliferation of adult T-cell leukemia cells.Retrovirology. 2011 Mar 17;8:19. doi: 10.1186/1742-4690-8-19. Retrovirology. 2011. PMID: 21414204 Free PMC article.
-
Adult T-cell leukemia-lymphoma as a viral disease: Subtypes based on viral aspects.Cancer Sci. 2021 May;112(5):1688-1694. doi: 10.1111/cas.14869. Epub 2021 Mar 30. Cancer Sci. 2021. PMID: 33630351 Free PMC article. Review.
Cited by
-
Humoral immune response to HTLV-1 basic leucine zipper factor (HBZ) in HTLV-1-infected individuals.Retrovirology. 2013 Feb 13;10:19. doi: 10.1186/1742-4690-10-19. Retrovirology. 2013. PMID: 23405908 Free PMC article.
-
Characterization of novel Bovine Leukemia Virus (BLV) antisense transcripts by deep sequencing reveals constitutive expression in tumors and transcriptional interaction with viral microRNAs.Retrovirology. 2016 May 3;13(1):33. doi: 10.1186/s12977-016-0267-8. Retrovirology. 2016. PMID: 27141823 Free PMC article.
-
New insight into the oncogenic mechanism of the retroviral oncoprotein Tax.Protein Cell. 2012 Aug;3(8):581-9. doi: 10.1007/s13238-012-2047-0. Epub 2012 Jul 21. Protein Cell. 2012. PMID: 22865346 Free PMC article. Review.
-
HDAC inhibitors Panobinostat and Romidepsin enhance tax transcription in HTLV-1-infected cell lines and freshly isolated patients' T-cells.Front Immunol. 2022 Aug 16;13:978800. doi: 10.3389/fimmu.2022.978800. eCollection 2022. Front Immunol. 2022. PMID: 36052071 Free PMC article.
-
Overview on HTLV-1 p12, p8, p30, p13: accomplices in persistent infection and viral pathogenesis.Front Microbiol. 2012 Dec 11;3:400. doi: 10.3389/fmicb.2012.00400. eCollection 2012. Front Microbiol. 2012. PMID: 23248621 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

