Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1
- PMID: 16407326
- PMCID: PMC1326240
- DOI: 10.1093/nar/gkj415
Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1
Abstract
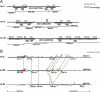
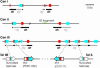
In Schizosaccharomyces pombe the RNAi machinery and proteins mediating heterochromatin formation regulate the transcription of non-coding centromeric repeats. These repeats share a high sequence similarity with telomere-linked helicase (tlh) genes, implying an ancestral relationship between the two types of elements and suggesting that transcription of the tlh genes might be regulated by the same factors as centromeric repeats. Indeed, we found that mutants lacking the histone methyltransferase Clr4, the Pcu4 cullin, Clr7 or Clr8, accumulate high levels of tlh forward and reverse transcripts. Mutations and conditions perturbing histone acetylation had similar effects further demonstrating that the tlh genes are normally repressed by heterochromatin. In contrast, mutations in the RNAi factors Dcr1, Ago1 or Rdp1 led only to a modest derepression of the tlh genes indicating an alternate pathway recruits heterochromatin components to telomeres. The telomere-binding protein Taz1 might be part of such a redundant pathway, tlh transcripts being present at low levels in Deltataz1 mutants and at higher levels in Deltataz1 Deltadcr1 double mutants. Surprisingly, the chromodomain protein Chp1, a component of the Ago1-containing RITS complex, contributes more to tlh repression than Ago1, indicating the repressive effects of Chp1 are partially independent of RITS. The tlh genes are found in the subtelomeric regions of several other fungi raising the intriguing possibility of conserved regulation and function.
Figures




Similar articles
-
Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres.Curr Biol. 2005 Oct 25;15(20):1808-19. doi: 10.1016/j.cub.2005.09.041. Curr Biol. 2005. PMID: 16243027
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
-
RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast.Mol Cell Biol. 2005 Mar;25(6):2331-46. doi: 10.1128/MCB.25.6.2331-2346.2005. Mol Cell Biol. 2005. PMID: 15743828 Free PMC article.
-
The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe.Genetics. 2005 Dec;171(4):1583-95. doi: 10.1534/genetics.105.048298. Epub 2005 Sep 12. Genetics. 2005. PMID: 16157682 Free PMC article.
-
On the connection between RNAi and heterochromatin at centromeres.Cold Spring Harb Symp Quant Biol. 2010;75:275-83. doi: 10.1101/sqb.2010.75.024. Epub 2011 Feb 2. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21289046 Review.
Cited by
-
Establishment of heterochromatin in domain-size-dependent bursts.Proc Natl Acad Sci U S A. 2021 Apr 13;118(15):e2022887118. doi: 10.1073/pnas.2022887118. Proc Natl Acad Sci U S A. 2021. PMID: 33827924 Free PMC article.
-
Characterization of genome-reduced fission yeast strains.Nucleic Acids Res. 2013 May 1;41(10):5382-99. doi: 10.1093/nar/gkt233. Epub 2013 Apr 5. Nucleic Acids Res. 2013. PMID: 23563150 Free PMC article.
-
Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe.PLoS One. 2012;7(11):e49151. doi: 10.1371/journal.pone.0049151. Epub 2012 Nov 2. PLoS One. 2012. PMID: 23133674 Free PMC article.
-
The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9.Epigenetics Chromatin. 2008 Nov 3;1(1):5. doi: 10.1186/1756-8935-1-5. Epigenetics Chromatin. 2008. PMID: 19014414 Free PMC article.
-
Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation.BMC Genomics. 2010 Jan 22;11:59. doi: 10.1186/1471-2164-11-59. BMC Genomics. 2010. PMID: 20096118 Free PMC article.
References
-
- Henikoff S., Dalal Y. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. - PubMed
-
- Allshire R. Centromere and kinetochore structure and function. In: Egel R., editor. The Molecular Biology of Schizosaccharomyce pombe. Berlin: Springer-Verlag; 2004. pp. 149–169.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

