A novel plant-specific family gene, ROOT PRIMORDIUM DEFECTIVE 1, is required for the maintenance of active cell proliferation
- PMID: 16407439
- PMCID: PMC1361326
- DOI: 10.1104/pp.105.074724
A novel plant-specific family gene, ROOT PRIMORDIUM DEFECTIVE 1, is required for the maintenance of active cell proliferation
Abstract
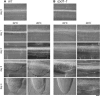
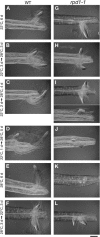
Hypocotyl segments of Arabidopsis (Arabidopsis thaliana) produce adventitious roots in response to exogenously supplied auxin. root primordium defective 1 (rpd1) is a temperature-sensitive mutant isolated on the basis of impairment in this phenomenon. This study describes further phenotypic analysis of the rpd1 mutant and isolation of the RPD1 gene. When adventitious root formation was induced from the rpd1 explants at the restrictive temperature, cell proliferation leading to root promordia formation was initiated at the same time as in wild-type explants. However, development of the root primordia was arrested thereafter in the mutant. Temperature-shift experiments indicated that RPD1 exerts its function before any visible sign of root primordium formation. The expression patterns of the auxin-responsive gene DR5:beta-glucuronidase and the cytodifferentiation marker gene SCARECROW suggest that the rpd1 mutation interferes with neither axis formation nor cellular patterning at the initial stage of root primordium development. Taken together with the effect of the rpd1 mutation on callus cell proliferation, these data imply a role for RPD1 in prearranging the maintenance of the active cell proliferation during root primordium development. Positional cloning of the RPD1 gene revealed that it encodes a member of a novel protein family specific to the plant kingdom. Disruption of the RPD1 gene by a T-DNA insertion caused embryogenesis arrest at the globular to transition stages. This phenotype is consistent with the hypothesized function of RPD1 in the maintenance of active cell proliferation.
Figures







Similar articles
-
Temperature-sensitive splicing defects in Arabidopsis mitochondria caused by mutations in the ROOT PRIMORDIUM DEFECTIVE 1 gene.Nucleic Acids Res. 2024 May 8;52(8):4575-4587. doi: 10.1093/nar/gkae072. Nucleic Acids Res. 2024. PMID: 38364869 Free PMC article.
-
Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana.Development. 2003 Dec;130(23):5637-47. doi: 10.1242/dev.00794. Epub 2003 Oct 1. Development. 2003. PMID: 14522871
-
Cytokinin receptors are required for normal development of auxin-transporting vascular tissues in the hypocotyl but not in adventitious roots.Plant Cell Physiol. 2006 Feb;47(2):234-43. doi: 10.1093/pcp/pci240. Epub 2005 Dec 15. Plant Cell Physiol. 2006. PMID: 16357038
-
Pivotal role of LBD16 in root and root-like organ initiation.Cell Mol Life Sci. 2018 Sep;75(18):3329-3338. doi: 10.1007/s00018-018-2861-5. Epub 2018 Jun 25. Cell Mol Life Sci. 2018. PMID: 29943076 Free PMC article. Review.
-
Arabidopsis embryogenesis. Radicle development(s).Curr Biol. 1995 Feb 1;5(2):110-2. doi: 10.1016/s0960-9822(95)00027-3. Curr Biol. 1995. PMID: 7743170 Review.
Cited by
-
A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing.Proc Natl Acad Sci U S A. 2009 Mar 17;106(11):4537-42. doi: 10.1073/pnas.0812503106. Epub 2009 Feb 26. Proc Natl Acad Sci U S A. 2009. PMID: 19251672 Free PMC article.
-
Elucidating circRNA-miRNA-mRNA competing endogenous regulatory RNA network during leaf rust pathogenesis in wheat (Triticum aestivum L.).Funct Integr Genomics. 2025 Jan 16;25(1):15. doi: 10.1007/s10142-024-01520-x. Funct Integr Genomics. 2025. PMID: 39815073
-
Rice FLOURY SHRUNKEN ENDOSPERM 5 Encodes a Putative Plant Organelle RNA Recognition Protein that Is Required for cis-Splicing of Mitochondrial nad4 Intron 1.Rice (N Y). 2021 Mar 10;14(1):29. doi: 10.1186/s12284-021-00463-2. Rice (N Y). 2021. PMID: 33689034 Free PMC article.
-
Temperature-sensitive splicing defects in Arabidopsis mitochondria caused by mutations in the ROOT PRIMORDIUM DEFECTIVE 1 gene.Nucleic Acids Res. 2024 May 8;52(8):4575-4587. doi: 10.1093/nar/gkae072. Nucleic Acids Res. 2024. PMID: 38364869 Free PMC article.
-
Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis.Sci Rep. 2019 Jul 22;9(1):10537. doi: 10.1038/s41598-019-47038-2. Sci Rep. 2019. PMID: 31332216 Free PMC article.
References
-
- Akama K, Shiraishi H, Ohta S, Nakamura K, Okada K, Shimura Y (1992) Efficient transformation of Arabidopsis thaliana: comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep 12: 7–11 - PubMed
-
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

