The fertilization-induced DNA replication factor MCM6 of maize shuttles between cytoplasm and nucleus, and is essential for plant growth and development
- PMID: 16407440
- PMCID: PMC1361320
- DOI: 10.1104/pp.105.074294
The fertilization-induced DNA replication factor MCM6 of maize shuttles between cytoplasm and nucleus, and is essential for plant growth and development
Abstract
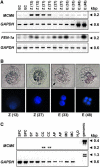
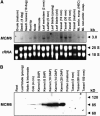
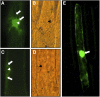
The eukaryotic genome is duplicated exactly once per cell division cycle. A strategy that limits every replication origin to a single initiation event is tightly regulated by a multiprotein complex, which involves at least 20 protein factors. A key player in this regulation is the evolutionary conserved hexameric MCM2-7 complex. From maize (Zea mays) zygotes, we have cloned MCM6 and characterized this essential gene in more detail. Shortly after fertilization, expression of ZmMCM6 is strongly induced. During progression of zygote and proembryo development, ZmMCM6 transcript amounts decrease and are low in vegetative tissues, where expression is restricted to tissues containing proliferating cells. The highest protein amounts are detectable about 6 to 20 d after fertilization in developing kernels. Subcellular localization studies revealed that MCM6 protein shuttles between cytoplasm and nucleoplasm in a cell cycle-dependent manner. ZmMCM6 is taken up by the nucleus during G1 phase and the highest protein levels were observed during late G1/S phase. ZmMCM6 is excluded from the nucleus during late S, G2, and mitosis. Transgenic maize was generated to overexpress and down-regulate ZmMCM6. Plants displaying minor antisense transcript amounts were reduced in size and did not develop cobs to maturity. Down-regulation of ZmMCM6 gene activity seems also to affect pollen development because antisense transgenes could not be propagated via pollen to wild-type plants. In summary, the transgenic data indicate that MCM6 is essential for both vegetative as well as reproductive growth and development in plants.
Figures



Similar articles
-
DiSUMO-like DSUL is required for nuclei positioning, cell specification and viability during female gametophyte maturation in maize.Development. 2010 Jan;137(2):333-45. doi: 10.1242/dev.035964. Development. 2010. PMID: 20040499
-
Maize DBF1-interactor protein 1 containing an R3H domain is a potential regulator of DBF1 activity in stress responses.Plant J. 2006 Jun;46(5):747-57. doi: 10.1111/j.1365-313X.2006.02742.x. Plant J. 2006. PMID: 16709191
-
TRANSPARENT LEAF AREA1 encodes a secreted proteolipid required for anther maturation, morphogenesis, and differentiation during leaf development in maize.Plant Cell. 2005 Mar;17(3):730-45. doi: 10.1105/tpc.104.028340. Epub 2005 Feb 10. Plant Cell. 2005. PMID: 15705951 Free PMC article.
-
Cell Biology of the Plant Nucleus.Annu Rev Plant Biol. 2017 Apr 28;68:139-172. doi: 10.1146/annurev-arplant-042916-041115. Epub 2017 Feb 22. Annu Rev Plant Biol. 2017. PMID: 28226231 Review.
-
Molecular basis of disease susceptibility in the Texas cytoplasm of maize.Plant Mol Biol. 1992 May;19(1):135-47. doi: 10.1007/BF00015611. Plant Mol Biol. 1992. PMID: 1600165 Review. No abstract available.
Cited by
-
Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize.Plant Cell. 2012 Dec;24(12):4974-91. doi: 10.1105/tpc.112.107169. Epub 2012 Dec 18. Plant Cell. 2012. PMID: 23250449 Free PMC article.
-
Plant MCM proteins: role in DNA replication and beyond.Plant Mol Biol. 2011 Dec;77(6):537-45. doi: 10.1007/s11103-011-9836-3. Epub 2011 Oct 25. Plant Mol Biol. 2011. PMID: 22038093 Review.
-
The plant cell cycle: Pre-Replication complex formation and controls.Genet Mol Biol. 2017;40(1 suppl 1):276-291. doi: 10.1590/1678-4685-GMB-2016-0118. Epub 2017 Mar 16. Genet Mol Biol. 2017. PMID: 28304073 Free PMC article.
-
Does Early Embryogenesis in Eudicots and Monocots Involve the Same Mechanism and Molecular Players?Plant Physiol. 2017 Jan;173(1):130-142. doi: 10.1104/pp.16.01406. Epub 2016 Dec 1. Plant Physiol. 2017. PMID: 27909044 Free PMC article. Review.
-
Transcriptomic Profiling Highlights Metabolic and Biosynthetic Pathways Involved in In Vitro Flowering in Anoectochilus roxburghii (Wall.) Lindl.Genes (Basel). 2025 Jan 24;16(2):132. doi: 10.3390/genes16020132. Genes (Basel). 2025. PMID: 40004461 Free PMC article.
References
-
- Bailis JM, Forsburg SL (2004) MCM proteins: DNA damage, mutagenesis and repair. Curr Opin Genet Dev 14: 17–21 - PubMed
-
- Bastida M, Puigdomènech P (2002) Specific expression of ZmPRL, the maize homolog of MCM7, during early embryogenesis. Plant Sci 162: 97–106
-
- Bogan JA, Natale DA, Depamphilis ML (2000) Initiation of eukaryotic DNA replication: conservative or liberal? J Cell Physiol 184: 139–150 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

