ETO2 coordinates cellular proliferation and differentiation during erythropoiesis
- PMID: 16407974
- PMCID: PMC1383517
- DOI: 10.1038/sj.emboj.7600934
ETO2 coordinates cellular proliferation and differentiation during erythropoiesis
Abstract
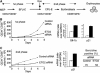
The passage from proliferation to terminal differentiation is critical for normal development and is often perturbed in malignancies. To define the molecular mechanisms that govern this process during erythropoiesis, we have used tagging/proteomics approaches and characterized protein complexes nucleated by TAL-1/SCL, a basic helix-loop-helix transcription factor that specifies the erythrocytic lineage. In addition to known TAL-1 partners, GATA-1, E2A, HEB, LMO2 and Ldb1, we identify the ETO2 repressor as a novel component recruited to TAL-1 complexes through interaction with E2A/HEB. Ectopic expression and siRNA knockdown experiments in hematopoietic progenitor cells show that ETO2 actively represses erythroid TAL-1 target genes and governs the expansion of erythroid progenitors. At the onset of erythroid differentiation, a change in the stoichiometry of ETO2 within the TAL-1 complex activates the expression of known erythroid-specific TAL-1 target genes and of Gfi-1b and p21(Cip), encoding two essential regulators of erythroid cell proliferation. These results suggest that the dynamics of ETO2 recruitment within nuclear complexes couple cell proliferation to cell differentiation and determine the onset of terminal erythroid maturation.
Figures



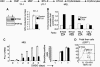

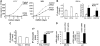
References
-
- Amsellem S, Ravet E, Fichelson S, Pflumio F, Dubart-Kupperschmitt A (2002) Maximal lentivirus-mediated gene transfer and sustained transgene expression in human hematopoietic primitive cells and their progeny. Mol Ther 6: 673–677 - PubMed
-
- Asari S, Sakamoto A, Okada S, Ohkubo Y, Arima M, Hatano M, Kuroda Y, Tokuhisa T (2005) Abnormal erythroid differentiation in neonatal bcl-6-deficient mice. Exp Hematol 33: 26–34 - PubMed
-
- Begley CG, Green AR (1999) The SCL gene: from case report to critical hematopoietic regulator. Blood 93: 2760–2770 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

