Enzymes: An integrated view of structure, dynamics and function
- PMID: 16409630
- PMCID: PMC1379655
- DOI: 10.1186/1475-2859-5-2
Enzymes: An integrated view of structure, dynamics and function
Abstract
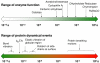
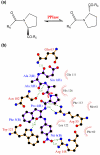
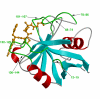
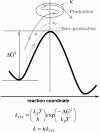
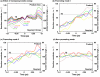
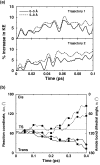
Microbes utilize enzymes to perform a variety of functions. Enzymes are biocatalysts working as highly efficient machines at the molecular level. In the past, enzymes have been viewed as static entities and their function has been explained on the basis of direct structural interactions between the enzyme and the substrate. A variety of experimental and computational techniques, however, continue to reveal that proteins are dynamically active machines, with various parts exhibiting internal motions at a wide range of time-scales. Increasing evidence also indicates that these internal protein motions play a role in promoting protein function such as enzyme catalysis. Moreover, the thermodynamical fluctuations of the solvent, surrounding the protein, have an impact on internal protein motions and, therefore, on enzyme function. In this review, we describe recent biochemical and theoretical investigations of internal protein dynamics linked to enzyme catalysis. In the enzyme cyclophilin A, investigations have lead to the discovery of a network of protein vibrations promoting catalysis. Cyclophilin A catalyzes peptidyl-prolyl cis/trans isomerization in a variety of peptide and protein substrates. Recent studies of cyclophilin A are discussed in detail and other enzymes (dihydrofolate reductase and liver alcohol dehydrogenase) where similar discoveries have been reported are also briefly discussed. The detailed characterization of the discovered networks indicates that protein dynamics plays a role in rate-enhancement achieved by enzymes. An integrated view of enzyme structure, dynamics and function have wide implications in understanding allosteric and co-operative effects, as well as protein engineering of more efficient enzymes and novel drug design.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

