Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin
- PMID: 16409631
- PMCID: PMC1360674
- DOI: 10.1186/1742-4690-3-4
Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin
Abstract
Background: The early events of the HIV-1 life cycle include entry of the viral core into target cell, assembly of the reverse transcription complex (RTCs) performing reverse transcription, its transformation into integration-competent complexes called pre-integration complexes (PICs), trafficking of complexes into the nucleus, and finally integration of the viral DNA into chromatin. Molecular details and temporal organization of these processes remain among the least investigated and most controversial problems in the biology of HIV.
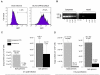
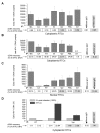
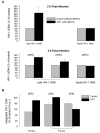
Results: To quantitatively evaluate maturation and nuclear translocation of the HIV-1 RTCs, nucleoprotein complexes isolated from the nucleus (nRTC) and cytoplasm (cRTC) of HeLa cells infected with MLV Env-pseudotyped HIV-1 were analyzed by real-time PCR. While most complexes completed reverse transcription in the cytoplasm, some got into the nucleus before completing DNA synthesis. The HIV-specific RNA complexes could get into the nucleus when reverse transcription was blocked by reverse transcriptase inhibitor, although nuclear import of RNA complexes was less efficient than of DNA-containing RTCs. Analysis of the RTC nuclear import in synchronized cells infected in the G2/M phase of the cell cycle showed enrichment in the nuclei of RTCs containing incomplete HIV-1 DNA compared to non-synchronized cells, where RTCs with complete reverse transcripts prevailed. Immunoprecipitation assays identified viral proteins IN, Vpr, MA, and cellular Ini1 and PML associated with both cRTCs and nRTCs, whereas CA was detected only in cRTCs and RT was diminished in nRTCs. Cytoplasmic maturation of the complexes was associated with increased immunoreactivity with anti-Vpr and anti-IN antibodies, and decreased reactivity with antibodies to RT. Both cRTCs and nRTCs carried out endogenous reverse transcription reaction in vitro. In contrast to cRTCs, in vitro completion of reverse transcription in nRTCs did not increase their integration into chromatin.
Conclusion: These results suggest that RTC maturation occurs predominantly in the cytoplasm. Immature RTCs containing RT and incomplete DNA can translocate into the nucleus during mitosis and complete reverse transcription, but are defective for integration.
Figures


References
-
- Heinzinger NK, Bukrinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci U S A. 1994;91:7311–7315. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

