Bioinformatic analysis of an unusual gene-enzyme relationship in the arginine biosynthetic pathway among marine gamma proteobacteria: implications concerning the formation of N-acetylated intermediates in prokaryotes
- PMID: 16409639
- PMCID: PMC1382215
- DOI: 10.1186/1471-2164-7-4
Bioinformatic analysis of an unusual gene-enzyme relationship in the arginine biosynthetic pathway among marine gamma proteobacteria: implications concerning the formation of N-acetylated intermediates in prokaryotes
Abstract
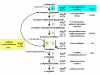
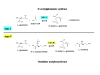
Background: The N-acetylation of L-glutamate is regarded as a universal metabolic strategy to commit glutamate towards arginine biosynthesis. Until recently, this reaction was thought to be catalyzed by either of two enzymes: (i) the classical N-acetylglutamate synthase (NAGS, gene argA) first characterized in Escherichia coli and Pseudomonas aeruginosa several decades ago and also present in vertebrates, or (ii) the bifunctional version of ornithine acetyltransferase (OAT, gene argJ) present in Bacteria, Archaea and many Eukaryotes. This paper focuses on a new and surprising aspect of glutamate acetylation. We recently showed that in Moritella abyssi and M. profunda, two marine gamma proteobacteria, the gene for the last enzyme in arginine biosynthesis (argH) is fused to a short sequence that corresponds to the C-terminal, N-acetyltransferase-encoding domain of NAGS and is able to complement an argA mutant of E. coli. Very recently, other authors identified in Mycobacterium tuberculosis an independent gene corresponding to this short C-terminal domain and coding for a new type of NAGS. We have investigated the two prokaryotic Domains for patterns of gene-enzyme relationships in the first committed step of arginine biosynthesis.
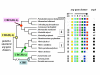
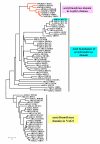
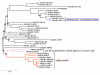
Results: The argH-A fusion, designated argH(A), and discovered in Moritella was found to be present in (and confined to) marine gamma proteobacteria of the Alteromonas- and Vibrio-like group. Most of them have a classical NAGS with the exception of Idiomarina loihiensis and Pseudoalteromonas haloplanktis which nevertheless can grow in the absence of arginine and therefore appear to rely on the arg(A) sequence for arginine biosynthesis. Screening prokaryotic genomes for virtual argH-X 'fusions' where X stands for a homologue of arg(A), we retrieved a large number of Bacteria and several Archaea, all of them devoid of a classical NAGS. In the case of Thermus thermophilus and Deinococcus radiodurans, the arg(A)-like sequence clusters with argH in an operon-like fashion. In this group of sequences, we find the short novel NAGS of the type identified in M. tuberculosis. Among these organisms, at least Thermus, Mycobacterium and Streptomyces species appear to rely on this short NAGS version for arginine biosynthesis.
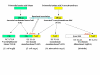
Conclusion: The gene-enzyme relationship for the first committed step of arginine biosynthesis should now be considered in a new perspective. In addition to bifunctional OAT, nature appears to implement at least three alternatives for the acetylation of glutamate. It is possible to propose evolutionary relationships between them starting from the same ancestral N-acetyltransferase domain. In M. tuberculosis and many other bacteria, this domain evolved as an independent enzyme, whereas it fused either with a carbamate kinase fold to give the classical NAGS (as in E. coli) or with argH as in marine gamma proteobacteria. Moreover, there is an urgent need to clarify the current nomenclature since the same gene name argA has been used to designate structurally different entities. Clarifying the confusion would help to prevent erroneous genomic annotation.
Figures

References
-
- Charlier D, Glansdorff N. Biosynthesis of arginine and polyamines. In: Curtiss R III (Editor in Chief), editor. EcoSal – Escherichia coli and Salmonella: Cellular and Molecular Biology Online. ASM Press, Washington, D. C.; Module 3.6.1.10; 2004. http://www.ecosal.org
-
- Shi D, Morizono H, Xiaolin Y, Roth L, Caldovic L, Allewell NM, Malamy MH, Tuchman M. Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria. J Biol Chem. 2005;280:14366–14369. doi: 10.1074/jbc.C500005200. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

