Unexpected NO-dependent DNA binding by the CooA homolog from Carboxydothermus hydrogenoformans
- PMID: 16410360
- PMCID: PMC1347970
- DOI: 10.1073/pnas.0505919103
Unexpected NO-dependent DNA binding by the CooA homolog from Carboxydothermus hydrogenoformans
Abstract
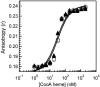
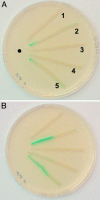
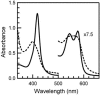
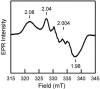
CooA, the CO-sensing heme protein from Rhodospirillum rubrum, regulates the expression of genes that encode a CO-oxidation system, allowing R. rubrum to use CO as a sole energy source. To better understand the gas-sensing regulation mechanism used by R. rubrum CooA and its homologs in other organisms, we characterized spectroscopically and functionally the Fe(II), Fe(II)-NO, and Fe(II)-CO forms of CooA from Carboxydothermus hydrogenoformans. Surprisingly, and unlike R. rubrum CooA, C. hydrogenoformans CooA binds NO to form a six-coordinate Fe(II)-NO heme that is active for DNA binding in vitro and in vivo. In contrast, R. rubrum CooA, which is exquisitely specific for CO, forms a five-coordinate Fe(II)-NO adduct that is inactive for DNA binding. Based on analyses of protein variants and temperature studies, NO-dependent DNA binding by C. hydrogenoformans CooA is proposed to result from a greater apparent stability of the six-coordinate Fe(II)-NO adduct at room temperature. Results from the present study strengthen the proposal that CO specificity in the CooA activation mechanism is based on the requirement for a small, neutral distal ligand, which in turn affects the relative positioning of the ligand-bound heme.
Figures
References
-
- Jain, R. & Chan, M. K. (2003) J. Biol. Inorg. Chem. 8, 1–11. - PubMed
-
- Gilles-Gonzalez, M.-A. & Gonzalez, G. (2005) J. Inorg. Biochem. 99, 1–22. - PubMed
-
- Roberts, G. P., Kerby, R. L., Youn, H. & Conrad, M. (2005) J. Inorg. Biochem. 99, 280–292. - PubMed
-
- Aono, S. (2003) Acc. Chem. Res. 36, 825–831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

