Photolabeling of cardiolipin binding subunits within bovine heart cytochrome c oxidase
- PMID: 16411750
- PMCID: PMC2561917
- DOI: 10.1021/bi050870z
Photolabeling of cardiolipin binding subunits within bovine heart cytochrome c oxidase
Abstract
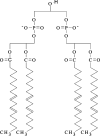
Subunits located near the cardiolipin binding sites of bovine heart cytochrome c oxidase (CcO) were identified by photolabeling with arylazido-cardiolipin analogues and detecting labeled subunits by reversed-phase HPLC and HPLC-electrospray ionization mass spectrometry. Two arylazido-containing cardiolipin analogues were synthesized: (1) 2-SAND-gly-CL with a nitrophenylazido group attached to the polar headgroup of cardiolipin (CL) via a linker containing a cleavable disulfide; (2) 2',2''-bis-(AzC12)-CL with two of the four fatty acid tails of cardiolipin replaced by 12-(N-4-azido-2-nitrophenyl) aminododecanoic acid. Both arylazido-CL derivatives were used to map the cardiolipin binding sites within two types of detergent-solubilized CcO: (1) intact 13-subunit CL-containing CcO (three to four molecules of endogenous CL remain bound per CcO monomer); (2) 11-subunit CL-free CcO (subunits VIa and VIb are missing because they dissociate during CL removal). Upon the basis of these photolabeling studies, we conclude that (1) subunits VIIa, VIIc, and possibly VIII are located near the two high-affinity cardiolipin binding sites, which are present in either form of CcO, and (2) subunit VIa is located adjacent to the lower affinity cardiolipin binding site, which is only present in the 13-subunit form of CcO. These data are consistent with the recent CcO crystal structure in which one cardiolipin is located near subunit VIIa and a second is located near subunit VIa (PDB ID code referenced in Tomitake, T. et al. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15304-15309). However, we propose that a third cardiolipin is bound between subunits VIIa and VIIc near the entrance to the D-channel. Cardiolipin bound at this location could potentially function as a proton antenna to facilitate proton entry into the D-channel. If true, it would explain the CcO requirement of bound cardiolipin for full electron transport activity.
Figures





References
-
- Kadenbach B, Jarausch J, Hartmann R, Merle P. Separtation of Mammalian Cytochrome c Oxidase into 13 Polypeptides by a Sodium Dodecyl Sulfate-Gel Electrophoresis Procedure. Anal. Biochem. 1983;129:517–521. - PubMed
-
- Ioannou PV, Golding BT. Cardiolipins: Their Chemistry and Biochemistry. J. Lipid Res. 1979;17:279–318. - PubMed
-
- Hoch FL. Cardiolipins and Biomembrane Function. Biochim. Biophys. Acta. 1992;1113:71–133. - PubMed
-
- Schlame M, Rua D, Greenberg ML. The Biosynthesis and Functional Role of Cardiolipin. Prog. Lipid Res. 2000;39:257–288. - PubMed
-
- Robinson NC. Functional Binding of Cardiolipin to Cytochromec Oxidase. J. Bioenerg. Biomembr. 1993;25:153–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

