Fluoride inhibition of enolase: crystal structure and thermodynamics
- PMID: 16411755
- PMCID: PMC2566932
- DOI: 10.1021/bi051558s
Fluoride inhibition of enolase: crystal structure and thermodynamics
Abstract
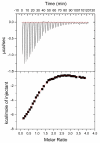
Enolase is a dimeric metal-activated metalloenzyme which uses two magnesium ions per subunit: the strongly bound conformational ion and the catalytic ion that binds to the enzyme-substrate complex inducing catalysis. The crystal structure of the human neuronal enolase-Mg2F2P(i) complex (enolase fluoride/phosphate inhibitory complex, EFPIC) determined at 1.36 A resolution shows that the combination of anions effectively mimics an intermediate state in catalysis. The phosphate ion binds in the same site as the phosphate group of the substrate/product, 2-phospho-D-glycerate/phosphoenolpyruvate, and induces binding of the catalytic Mg2+ ion. One fluoride ion bridges the structural and catalytic magnesium ions while the other interacts with the structural magnesium ion and the ammonio groups of Lys 342 and Lys 393. These fluoride ion positions correspond closely to the positions of the oxygen atoms of the substrate's carboxylate moiety. To relate structural changes resulting from fluoride, phosphate, and magnesium ions binding to those that are induced by phosphate and magnesium ions alone, we also determined the structure of the human neuronal enolase-Mg2P(i) complex (enolase phosphate inhibitory complex, EPIC) at 1.92 A resolution. It shows the closed conformation in one subunit and a mixture of open and semiclosed conformations in the other. The EPFIC dimer is essentially symmetric while the EPIC dimer is asymmetric. Isothermal titration calorimetry data confirmed binding of four fluoride ions per dimer and yielded Kb values of 7.5 x 10(5) +/- 1.3 x 10(5), 1.2 x 10(5) +/- 0.2 x 10(5), 8.6 x 10(4) +/- 1.6 x 10(4), and 1.6 x 10(4) +/- 0.7 x 10(4) M(-1). The different binding constants indicate negative cooperativity between the subunits; the asymmetry of EPIC supports such an interpretation.
Figures
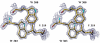





References
-
- Warburg O, Christian W. Isolierung und Kristallisation des Garungsferments Enolase. Biochem. Z. 1941;310:384–421.
-
- Brewer JM. Yeast enolase: mechanism of activation by metal ions. CRC Crit. Rev. Biochem. 1981;11:209–254. - PubMed
-
- Larsen TM, Wedekind JE, Rayment I, Reed GH. A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution. Biochemistry. 1996;35:4349–58. - PubMed
-
- Zhang E, Brewer JM, Minor W, Carreira LA, Lebioda L. Mechanism of enolase: the crystal structure of asymmetric dimer enolase- 2-phospho-D-glycerate/enolase-phosphoenolpyruvate at 2.0 Å resolution. Biochemistry. 1997;36:12526–12534. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

