Crystal structure of inactivated Thermotoga maritima invertase in complex with the trisaccharide substrate raffinose
- PMID: 16411890
- PMCID: PMC1462702
- DOI: 10.1042/BJ20051936
Crystal structure of inactivated Thermotoga maritima invertase in complex with the trisaccharide substrate raffinose
Abstract
Thermotoga maritima invertase (beta-fructosidase), a member of the glycoside hydrolase family GH-32, readily releases beta-D-fructose from sucrose, raffinose and fructan polymers such as inulin. These carbohydrates represent major carbon and energy sources for prokaryotes and eukaryotes. The invertase cleaves beta-fructopyranosidic linkages by a double-displacement mechanism, which involves a nucleophilic aspartate and a catalytic glutamic acid acting as a general acid/base. The three-dimensional structure of invertase shows a bimodular enzyme with a five bladed beta-propeller catalytic domain linked to a beta-sandwich of unknown function. In the present study we report the crystal structure of the inactivated invertase in interaction with the natural substrate molecule alpha-D-galactopyranosyl-(1,6)-alpha-D-glucopyranosyl-beta-D-fructofuranoside (raffinose) at 1.87 A (1 A=0.1 nm) resolution. The structural analysis of the complex reveals the presence of three binding-subsites, which explains why T. maritima invertase exhibits a higher affinity for raffinose than sucrose, but a lower catalytic efficiency with raffinose as substrate than with sucrose.
Figures

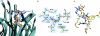
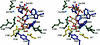
References
-
- Ritsema T., Smeekens S. Fructans: beneficial for plants and humans. Curr. Opin. Plant Biol. 2003;6:223–230. - PubMed
-
- Sturm A., Tang G. Q. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999;4:401–407. - PubMed
-
- Koshland D. E., Jr, Stein S. S. Correlation of bond breaking with enzyme specificity; cleavage point of invertase. J. Biol. Chem. 1954;208:139–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

