Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated
- PMID: 16413043
- PMCID: PMC7111823
- DOI: 10.1016/j.virol.2005.11.044
Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated
Abstract
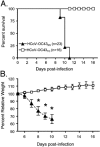
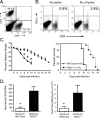
The human coronavirus HCoV-OC43 causes a significant fraction of upper respiratory tract infections. Most coronaviruses show a strong species specificity, although the SARS-Coronavirus crossed species from palm civet cats to infect humans. Similarly, HCoV-OC43, likely a member of the same coronavirus group as SARS-CoV, readily crossed the species barrier as evidenced by its rapid adaptation to the murine brain [McIntosh, K., Becker, W.B., Chanock, R.M., 1967. Growth in suckling-mouse brain of "IBV-like" viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci U.S.A. 58, 2268-73]. Herein, we investigated two consequences of this plasticity in species tropism. First, we showed that HCoV-OC43 was able to infect cells from a large number of mammalian species. Second, we showed that virus that was passed exclusively in suckling mouse brains was highly virulent and caused a uniformly fatal encephalitis in adult mice. The surface glycoprotein is a major virulence factor in most coronavirus infections. We identified three changes in the HCoV-OC43 surface glycoprotein that correlated with enhanced neurovirulence in mice; these were located in the domain of the protein responsible for binding to host cells. These data suggest that some coronaviruses, including HCoV-OC43 and SARS-CoV, readily adapt to growth in cells from heterologous species. This adaptability has facilitated the isolation of HCoV-OC43 viral variants with markedly differing abilities to infect animals and tissue culture cells.
Figures








References
-
- Barthold S.W., de Souza M.S., Smith A.L. Susceptibility of laboratory mice to intranasal and contact infection with coronaviruses of other species. Lab. Anim. Sci. 1990;40(5):481–485. - PubMed
-
- Bergmann C.C., Altman J.D., Hinton D., Stohlman S.A. Inverted immunodominance and impaired cytolytic function of CD8+T cells during viral persistence in the central nervous system. J. Immunol. 1999;163:3379–3387. - PubMed
-
- Binder G.K., Griffin D.E. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science. 2001;293(5528):303–306. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

