West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection
- PMID: 16415006
- PMCID: PMC1346927
- DOI: 10.1128/JVI.80.3.1290-1301.2006
West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection
Abstract
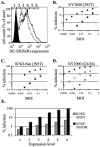
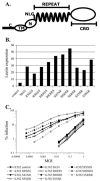
The C-type lectins DC-SIGN and DC-SIGNR bind mannose-rich glycans with high affinity. In vitro, cells expressing these attachment factors efficiently capture, and are infected by, a diverse array of appropriately glycosylated pathogens, including dengue virus. In this study, we investigated whether these lectins could enhance cellular infection by West Nile virus (WNV), a mosquito-borne flavivirus related to dengue virus. We discovered that DC-SIGNR promoted WNV infection much more efficiently than did DC-SIGN, particularly when the virus was grown in human cell types. The presence of a single N-linked glycosylation site on either the prM or E glycoprotein of WNV was sufficient to allow DC-SIGNR-mediated infection, demonstrating that uncleaved prM protein present on a flavivirus virion can influence viral tropism under certain circumstances. Preferential utilization of DC-SIGNR was a specific property conferred by the WNV envelope glycoproteins. Chimeras between DC-SIGN and DC-SIGNR demonstrated that the ability of DC-SIGNR to promote WNV infection maps to its carbohydrate recognition domain. WNV virions and subviral particles bound to DC-SIGNR with much greater affinity than DC-SIGN. We believe this is the first report of a pathogen interacting more efficiently with DC-SIGNR than with DC-SIGN. Our results should lead to the discovery of new mechanisms by which these well-studied lectins discriminate among ligands.
Figures




References
-
- Baribaud, F., R. W. Doms, and S. Pohlmann. 2002. The role of DC-SIGN and DC-SIGNR in HIV and Ebola virus infection: can potential therapeutics block virus transmission and dissemination? Expert Opin. Ther. Targets 6:423-431. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007324/AI/NIAID NIH HHS/United States
- U54 AI 57168/AI/NIAID NIH HHS/United States
- T32 AI 07324-13/AI/NIAID NIH HHS/United States
- T32 GM 007229/GM/NIGMS NIH HHS/United States
- T32 AI007632/AI/NIAID NIH HHS/United States
- T32 GM007229/GM/NIGMS NIH HHS/United States
- F31 RR005074/RR/NCRR NIH HHS/United States
- T32 AI 07632/AI/NIAID NIH HHS/United States
- U54 AI057168/AI/NIAID NIH HHS/United States
- AI 50469/AI/NIAID NIH HHS/United States
- F31 RR 05074/RR/NCRR NIH HHS/United States
- R01 AI050469/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

