Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production
- PMID: 16415015
- PMCID: PMC1346933
- DOI: 10.1128/JVI.80.3.1376-1384.2006
Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production
Abstract
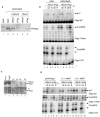
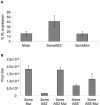
Posttranscriptional gene silencing allows sequence-specific control of gene expression. Specificity is guaranteed by small antisense RNAs such as microRNAs (miRNAs) or small interfering RNAs (siRNAs). Functional miRNAs derive from longer double-stranded RNA (dsRNA) molecules that are cleaved to pre-miRNAs in the nucleus and are transported by exportin 5 (Exp 5) to the cytoplasm. Adenovirus-infected cells express virus-associated (VA) RNAs, which are dsRNA molecules similar in structure to pre-miRNAs. VA RNAs are also transported by Exp 5 to the cytoplasm, where they accumulate. Here we show that small RNAs derived from VA RNAs (svaRNAs), similar to miRNAs, can be found in adenovirus-infected cells. VA RNA processing to svaRNAs requires neither viral replication nor viral protein expression, as evidenced by the fact that svaRNA accumulation can be detected in cells transfected with VA sequences. svaRNAs are efficiently bound by Argonaute 2, the endonuclease of the RNA-induced silencing complex, and behave as functional siRNAs, in that they inhibit the expression of reporter genes with complementary sequences. Blocking svaRNA-mediated inhibition affects efficient adenovirus production, indicating that svaRNAs are required for virus viability. Thus, svaRNA-mediated silencing could represent a novel mechanism used by adenoviruses to control cellular or viral gene expression.
Figures





References
-
- Bennasser, Y., S. Y. Le, M. Benkirane, and K. T. Jeang. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607-619. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

