Role of helix P of the human cytomegalovirus DNA polymerase in resistance and hypersusceptibility to the antiviral drug foscarnet
- PMID: 16415021
- PMCID: PMC1346920
- DOI: 10.1128/JVI.80.3.1440-1450.2006
Role of helix P of the human cytomegalovirus DNA polymerase in resistance and hypersusceptibility to the antiviral drug foscarnet
Abstract
Mutations in the human cytomegalovirus DNA polymerase (UL54) can not only decrease but also increase susceptibility to the pyrophosphate (PP(i)) analogue foscarnet. The proximity of L802M, which confers resistance, and K805Q, which confers hypersusceptibility, suggests a possible unifying mechanism that affects drug susceptibility in one direction or the other. We found that the polymerase activities of L802M- and K805Q-containing mutant enzymes were literally indistinguishable from that of wild-type UL54; however, susceptibility to foscarnet was decreased or increased, respectively. A comparison with the crystal structure model of the related RB69 polymerase suggests that L802 and K805 are located in the conserved alpha-helix P that is implicated in nucleotide binding. Although L802 and K805 do not appear to make direct contacts with the incoming nucleotide, it is conceivable that changes at these residues could exert their effects through the adjacent, highly conserved amino acids Q807 and/or K811. Our data show that a K811A substitution in UL54 causes reductions in rates of nucleotide incorporation. The activity of the Q807A mutant is only marginally affected, while this enzyme shows relatively high levels of resistance to foscarnet. Based on these data, we suggest that L802M exerts its effects through subtle structural changes in alpha-helix P that affect the precise positioning of Q807 and, in turn, its presumptive involvement in binding of foscarnet. In contrast, the removal of a positive charge associated with the K805Q change may facilitate access or increase affinity to the adjacent Q807.
Figures



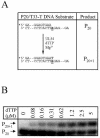
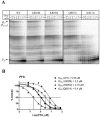
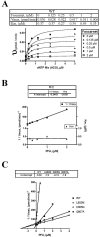
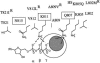
References
-
- Boosalis, M. S., J. Petruska, and M. F. Goodman. 1987. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 262:14689-14696. - PubMed
-
- Chou, S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. - PubMed
-
- Chou, S. W. 2001. Cytomegalovirus drug resistance and clinical implications. Transpl. Infect. Dis. 3(Suppl. 2):20-24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

