The cellular localization pattern of Varicella-Zoster virus ORF29p is influenced by proteasome-mediated degradation
- PMID: 16415026
- PMCID: PMC1346923
- DOI: 10.1128/JVI.80.3.1497-1512.2006
The cellular localization pattern of Varicella-Zoster virus ORF29p is influenced by proteasome-mediated degradation
Abstract
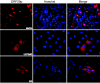
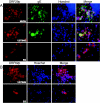
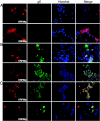
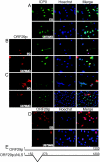
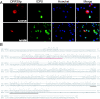
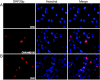
Varicella-zoster virus (VZV) open reading frame 29 (ORF29) encodes a single-stranded DNA binding protein. During lytic infection, ORF29p is localized primarily to infected-cell nuclei, whereas during latency it appears in the cytoplasm of infected neurons. Following reactivation, ORF29p accumulates in the nucleus. In this report, we analyze the cellular localization patterns of ORF29p during VZV infection and during autonomous expression. Our results demonstrate that ORF29p is excluded from the nucleus in a cell-type-specific manner and that its cellular localization pattern may be altered by subsequent expression of VZV ORF61p or herpes simplex virus type 1 ICP0. In these cases, ORF61p and ICP0 induce nuclear accumulation of ORF29p in cell lines where it normally remains cytoplasmic. One cellular system utilized by ICP0 to influence protein abundance is the proteasome degradation pathway. Inhibition of the 26S proteasome, but not heat shock treatment, resulted in accumulation of ORF29p in the nucleus, similar to the effect of ICP0 expression. Immunofluorescence microscopy and pulse-chase experiments reveal that stabilization of ORF29p correlates with its nuclear accumulation and is dependent on a functional nuclear localization signal. ORF29p nuclear translocation in cultured enteric neurons and cells derived from an astrocytoma is reversible, as the protein's distribution and stability revert to the previous states when the proteasomal activity is restored. Thus, stabilization of ORF29p leads to its nuclear accumulation. Although proteasome inhibition induces ORF29p nuclear accumulation, this is not sufficient to reactivate latent VZV or target the immediate-early protein ORF62p to the nucleus in cultured guinea pig enteric neurons.
Figures






Similar articles
-
Posttranslational modification and cell type-specific degradation of varicella-zoster virus ORF29p.J Virol. 2006 Nov;80(21):10836-46. doi: 10.1128/JVI.00966-06. Epub 2006 Sep 6. J Virol. 2006. PMID: 16956951 Free PMC article.
-
Dissection of a novel nuclear localization signal in open reading frame 29 of varicella-zoster virus.J Virol. 2005 Oct;79(20):13070-81. doi: 10.1128/JVI.79.20.13070-13081.2005. J Virol. 2005. PMID: 16189009 Free PMC article.
-
Nuclear import of the varicella-zoster virus latency-associated protein ORF63 in primary neurons requires expression of the lytic protein ORF61 and occurs in a proteasome-dependent manner.J Virol. 2008 Sep;82(17):8673-86. doi: 10.1128/JVI.00685-08. Epub 2008 Jun 18. J Virol. 2008. PMID: 18562514 Free PMC article.
-
Lessons to be learned from varicella-zoster virus.Vet Microbiol. 1996 Nov;53(1-2):55-66. doi: 10.1016/s0378-1135(96)01234-5. Vet Microbiol. 1996. PMID: 9010998 Review.
-
Intracellular synthesis of varicella-zoster virus.J Infect Dis. 1992 Aug;166 Suppl 1:S7-12. doi: 10.1093/infdis/166.supplement_1.s7. J Infect Dis. 1992. PMID: 1320653 Review.
Cited by
-
ORF7 of Varicella-Zoster Virus Is Required for Viral Cytoplasmic Envelopment in Differentiated Neuronal Cells.J Virol. 2017 May 26;91(12):e00127-17. doi: 10.1128/JVI.00127-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28356523 Free PMC article.
-
BAG3, a host cochaperone, facilitates varicella-zoster virus replication.J Virol. 2007 Jul;81(14):7491-503. doi: 10.1128/JVI.00442-07. Epub 2007 May 2. J Virol. 2007. PMID: 17475647 Free PMC article.
-
Infected peripheral blood mononuclear cells transmit latent varicella zoster virus infection to the guinea pig enteric nervous system.J Neurovirol. 2014 Oct;20(5):442-56. doi: 10.1007/s13365-014-0259-1. Epub 2014 Jun 26. J Neurovirol. 2014. PMID: 24965252 Free PMC article.
-
Proteasome inhibitors induce apoptosis of prostate cancer cells by inducing nuclear translocation of IkappaBalpha.Arch Biochem Biophys. 2008 Jul 15;475(2):156-63. doi: 10.1016/j.abb.2008.04.026. Epub 2008 Apr 29. Arch Biochem Biophys. 2008. PMID: 18468507 Free PMC article.
-
Sequencing and characterization of Varicella-zoster virus vaccine strain SuduVax.Virol J. 2011 Dec 16;8:547. doi: 10.1186/1743-422X-8-547. Virol J. 2011. PMID: 22176950 Free PMC article.
References
-
- Assouline, J. G., M. J. Levin, E. O. Major, B. Forghani, S. E. Straus, and J. M. Ostrove. 1990. Varicella-zoster virus infection of human astrocytes, Schwann cells, and neurons. Virology 179:834-844. - PubMed
-
- Boucaud, D., H. Yoshitake, J. Hay, and W. Ruyechan. 1998. The varicella-zoster virus (VZV) open-reading frame 29 protein acts as a modulator of a late VZV gene promoter. J Infect. Dis. 178(Suppl. 1):S34-S38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

