High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain
- PMID: 16415027
- PMCID: PMC1346936
- DOI: 10.1128/JVI.80.3.1513-1523.2006
High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain
Abstract
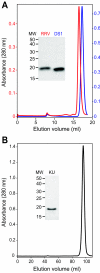
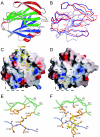
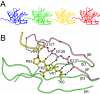
The most intensively studied rotavirus strains initially attach to cells when the "heads" of their protruding spikes bind cell surface sialic acid. Rotavirus strains that cause disease in humans do not bind this ligand. The structure of the sialic acid binding head (the VP8* core) from the simian rotavirus strain RRV has been reported, and neutralization epitopes have been mapped onto its surface. We report here a 1.6-A resolution crystal structure of the equivalent domain from the sialic acid-independent rotavirus strain DS-1, which causes gastroenteritis in humans. Although the RRV and DS-1 VP8* cores differ functionally, they share the same galectin-like fold. Differences between the RRV and DS-1 VP8* cores in the region that corresponds to the RRV sialic acid binding site make it unlikely that DS-1 VP8* binds an alternative carbohydrate ligand in this location. In the crystals, a surface cleft on each DS-1 VP8* core binds N-terminal residues from a neighboring molecule. This cleft may function as a ligand binding site during rotavirus replication. We also report an escape mutant analysis, which allows the mapping of heterotypic neutralizing epitopes recognized by human monoclonal antibodies onto the surface of the VP8* core. The distribution of escape mutations on the DS-1 VP8* core indicates that neutralizing antibodies that recognize VP8* of human rotavirus strains may bind a conformation of the spike that differs from those observed to date.
Figures
References
-
- Bartels, C., T.-H. Xia, M. Billeter, P. Guntert, and K. Wuthrich. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 5:1-10. - PubMed
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. - PubMed
-
- Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

