Molecular dynamics studies of the archaeal translocon
- PMID: 16415058
- PMCID: PMC1403164
- DOI: 10.1529/biophysj.105.075291
Molecular dynamics studies of the archaeal translocon
Abstract
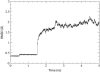
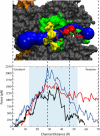
The translocon is a protein-conducting channel conserved over all domains of life that serves to translocate proteins across or into membranes. Although this channel has been well studied for many years, the recent discovery of a high-resolution crystal structure opens up new avenues of exploration. Taking advantage of this, we performed molecular dynamics simulations of the translocon in a fully solvated lipid bilayer, examining the translocation abilities of monomeric SecYEbeta by forcing two helices comprised of different amino acid sequences to cross the channel. The simulations revealed that the so-called plug of SecYEbeta swings open during translocation, closing thereafter. Likewise, it was established that the so-called pore ring region of SecYEbeta forms an elastic, yet tight, seal around the translocating oligopeptides. The closed state of the channel was found to block permeation of all ions and water molecules; in the open state, ions were blocked. Our results suggest that the SecYEbeta monomer is capable of forming an active channel.
Figures
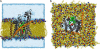

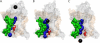

Similar articles
-
Simulations of a protein translocation pore: SecY.Biochemistry. 2006 Oct 31;45(43):13018-24. doi: 10.1021/bi061013d. Biochemistry. 2006. PMID: 17059218
-
Theoretical studies of the M2 transmembrane segment of the glycine receptor: models of the open pore structure and current-voltage characteristics.Biophys J. 2005 Sep;89(3):1669-80. doi: 10.1529/biophysj.105.060368. Epub 2005 Jun 10. Biophys J. 2005. PMID: 15951389 Free PMC article.
-
An energy-efficient gating mechanism in the acetylcholine receptor channel suggested by molecular and Brownian dynamics.Biophys J. 2006 Feb 1;90(3):799-810. doi: 10.1529/biophysj.105.067868. Epub 2005 Nov 11. Biophys J. 2006. PMID: 16284265 Free PMC article.
-
X-ray structure of a protein-conducting channel.Nature. 2004 Jan 1;427(6969):36-44. doi: 10.1038/nature02218. Epub 2003 Dec 3. Nature. 2004. PMID: 14661030
-
Mechanosensitive channels in archaea.Cell Biochem Biophys. 2001;34(3):349-81. doi: 10.1385/CBB:34:3:349. Cell Biochem Biophys. 2001. PMID: 11898861 Review.
Cited by
-
The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum.Int J Mol Sci. 2021 Dec 23;23(1):143. doi: 10.3390/ijms23010143. Int J Mol Sci. 2021. PMID: 35008565 Free PMC article. Review.
-
Long-timescale dynamics and regulation of Sec-facilitated protein translocation.Cell Rep. 2012 Oct 25;2(4):927-37. doi: 10.1016/j.celrep.2012.08.039. Epub 2012 Oct 19. Cell Rep. 2012. PMID: 23084746 Free PMC article.
-
Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA.EMBO J. 2007 Apr 18;26(8):1995-2004. doi: 10.1038/sj.emboj.7601661. Epub 2007 Mar 29. EMBO J. 2007. PMID: 17396152 Free PMC article.
-
Structural determinants of a permeation barrier of the SecYEG translocon in the active state.Phys Chem Chem Phys. 2021 Nov 24;23(45):25830-25840. doi: 10.1039/d1cp02702f. Phys Chem Chem Phys. 2021. PMID: 34762087 Free PMC article.
-
Amino-acid solvation structure in transmembrane helices from molecular dynamics simulations.Biophys J. 2006 Dec 15;91(12):4450-63. doi: 10.1529/biophysj.106.092767. Epub 2006 Sep 29. Biophys J. 2006. PMID: 17012325 Free PMC article.
References
-
- Matlack, K. E. S., W. Mothes, and T. A. Rapoport. 1998. Protein translocation: tunnel vision. Cell. 92:381–390. - PubMed
-
- Veenendaal, A. K. J., C. van der Does, and A. J. M. Driessen. 2004. The protein-conducting channel SecYEG. Biochim. Biophys. Acta. 1694:81–95. - PubMed
-
- Holland, I. B. 2004. Translocation of bacterial proteins—an overview. Biochim. Biophys. Acta. 1694:5–16. - PubMed
-
- Eichler, J. 2000. Archaeal protein translocation: crossing membranes in the third domain of life. Eur. J. Biochem. 267:3402–3412. - PubMed
-
- van den Berg, B., W. M. Clemons Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2004. X-ray structure of a protein-conducting channel. Nature. 427:36–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

