Local structural preferences and dynamics restrictions in the urea-denatured state of SUMO-1: NMR characterization
- PMID: 16415059
- PMCID: PMC1403170
- DOI: 10.1529/biophysj.105.071746
Local structural preferences and dynamics restrictions in the urea-denatured state of SUMO-1: NMR characterization
Abstract
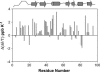
We have investigated by multidimensional NMR the structural and dynamic characteristics of the urea-denatured state of activated SUMO-1, a 97-residue protein belonging to the growing family of ubiquitin-like proteins involved in post-translational modifications. Complete backbone amide and 15N resonance assignments were obtained in the denatured state by using HNN and HN(C)N experiments. These enabled other proton assignments from TOCSY-HSQC spectra. Secondary Halpha chemical shifts and 1H-1H NOE indicate that the protein chain in the denatured state has structural preferences in the broad beta-domain for many residues. Several of these are seen to populate the (phi,psi) space belonging to polyproline II structure. Although there is no evidence for any persistent structures, many contiguous stretches of three or more residues exhibit structural propensities suggesting possibilities of short-range transient structure formation. The hetero-nuclear 1H-15N NOEs are extremely weak for most residues, except for a few at the C-terminal, and the 15N relaxation rates show sequence-wise variation. Some of the regions of slow motions coincide with those of structural preferences and these are interspersed by highly flexible residues. The implications of these observations for the early folding events starting from the urea-denatured state of activated SUMO-1 have been discussed.
Figures






Similar articles
-
NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding.Biochemistry. 2001 Mar 27;40(12):3561-71. doi: 10.1021/bi002776i. Biochemistry. 2001. PMID: 11297422
-
Residue-level NMR view of the urea-driven equilibrium folding transition of SUMO-1 (1-97): native preferences do not increase monotonously.J Mol Biol. 2006 Aug 4;361(1):180-94. doi: 10.1016/j.jmb.2006.06.003. Epub 2006 Jun 19. J Mol Biol. 2006. PMID: 16824543
-
1H, 15N, 13C resonance assignment of folded and 8 M urea-denatured state of SUMO from Drosophila melanogaster.Biomol NMR Assign. 2008 Jun;2(1):13-5. doi: 10.1007/s12104-007-9072-6. Epub 2007 Dec 4. Biomol NMR Assign. 2008. PMID: 19636913
-
Probing residual structure and backbone dynamics on the milli- to picosecond timescale in a urea-denatured fibronectin type III domain.J Mol Biol. 1999 Feb 19;286(2):579-92. doi: 10.1006/jmbi.1998.2479. J Mol Biol. 1999. PMID: 9973572
-
Structural and dynamic characterization of an unfolded state of poplar apo-plastocyanin formed under nondenaturing conditions.Protein Sci. 2001 May;10(5):1056-66. doi: 10.1110/ps.00601. Protein Sci. 2001. PMID: 11316886 Free PMC article.
Cited by
-
Guanidine-HCl dependent structural unfolding of M-crystallin: fluctuating native state like topologies and intermolecular association.PLoS One. 2012;7(12):e42948. doi: 10.1371/journal.pone.0042948. Epub 2012 Dec 17. PLoS One. 2012. PMID: 23284604 Free PMC article.
-
NMR insights into folding and self-association of Plasmodium falciparum P2.PLoS One. 2012;7(5):e36279. doi: 10.1371/journal.pone.0036279. Epub 2012 May 2. PLoS One. 2012. PMID: 22567147 Free PMC article.
-
Cooperative formation of native-like tertiary contacts in the ensemble of unfolded states of a four-helix protein.Proc Natl Acad Sci U S A. 2010 Jul 27;107(30):13306-11. doi: 10.1073/pnas.1003004107. Epub 2010 Jul 12. Proc Natl Acad Sci U S A. 2010. PMID: 20624986 Free PMC article.
-
Effect of an Imposed Contact on Secondary Structure in the Denatured State of Yeast Iso-1-cytochrome c.Biochemistry. 2017 Dec 26;56(51):6662-6676. doi: 10.1021/acs.biochem.7b01002. Epub 2017 Dec 8. Biochemistry. 2017. PMID: 29148740 Free PMC article.
-
NMR characterization of the near native and unfolded states of the PTB domain of Dok1: alternate conformations and residual clusters.PLoS One. 2014 Feb 28;9(2):e90557. doi: 10.1371/journal.pone.0090557. eCollection 2014. PLoS One. 2014. PMID: 24587391 Free PMC article.
References
-
- Religa, T. L., J. S. Markson, U. Mayor, S. M. Freund, and A. R. Fersht. 2005. Solution structure of a protein denatured state and folding intermediate. Nature. 437:1053–1056. - PubMed
-
- Bhavesh, N. S., R. Sinha, P. M. Mohan, and R. V. Hosur. 2003. NMR elucidation of early folding hierarchy in HIV-1 protease. J. Biol. Chem. 278:19980–19985. - PubMed
-
- Plotkin, S. S., and J. N. Onuchic. 2002. Understanding protein folding with energy landscape theory. Part I: Basic concepts. Q. Rev. Biophys. 35:111–167. - PubMed
-
- Plotkin, S. S., and J. N. Onuchic. 2002. Understanding protein folding with energy landscape theory. Part II: Quantitative aspects. Q. Rev. Biophys. 35:205–286. - PubMed
-
- Chan, H. S., and K. A. Dill. 1998. Protein folding in the landscape perspective: chevron plots and non-Arrhenius kinetics. Proteins. 30:2–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

