The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication
- PMID: 16415207
- PMCID: PMC1356546
- DOI: 10.1105/tpc.105.037309
The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication
Abstract
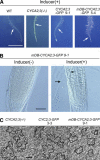
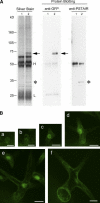
Plant cells frequently undergo endoreduplication, a process in which chromosomal DNA is successively duplicated in the absence of mitosis. It has been proposed that endoreduplication is regulated at its entry by mitotic cyclin-dependent kinase activity. However, the regulatory mechanisms for its termination remain unclear, although plants tightly control the ploidy level in each cell type. In the process of searching for regulatory factors of endoreduplication, the promoter of an Arabidopsis thaliana cyclin A gene, CYCA2;3, was revealed to be active in developing trichomes during the termination period of endoreduplication as well as in proliferating tissues. Taking advantage of the situation that plants encode highly redundant cyclin A genes, we were able to perform functional dissection of CYCA2;3 using null mutant alleles. Null mutations of CYCA2;3 semidominantly promoted endocycles and increased the ploidy levels achieved in mature organs, but they did not significantly affect the proportion of cells that underwent endoreduplication. Consistent with this result, expression of the CYCA2;3-green fluorescent protein fusion protein restrained endocycles in a dose-dependent manner. Moreover, a mutation in the destruction box of CYCA2;3 stabilized the fusion protein in the nuclei and enhanced the restraint. We conclude that CYCA2;3 negatively regulates endocycles and acts as a key regulator of ploidy levels in Arabidopsis endoreduplication.
Figures

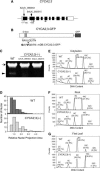
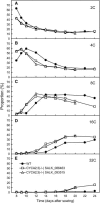

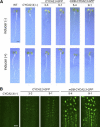
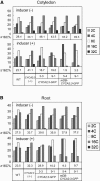

References
-
- Boudolf, V., Vlieghe, K., Beemster, G.T.S., Magyar, Z., Torres Acosta, J.A., Maes, S., Van Der Scheren, E., Inzé, D., and De Veylder, L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692. - PMC - PubMed
-
- Burssens, S., de Almeida Engler, J., Beeckman, T., Richard, C., Shaul, O., Ferreira, P., Van Montagu, M., and Inzé, D. (2000). Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211 623–631. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

