Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers
- PMID: 16417408
- PMCID: PMC1334226
- DOI: 10.1371/journal.pmed.0030047
Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers
Abstract
Background: Inadequate oxygen (hypoxia) triggers a multifaceted cellular response that has important roles in normal physiology and in many human diseases. A transcription factor, hypoxia-inducible factor (HIF), plays a central role in the hypoxia response; its activity is regulated by the oxygen-dependent degradation of the HIF-1alpha protein. Despite the ubiquity and importance of hypoxia responses, little is known about the variation in the global transcriptional response to hypoxia among different cell types or how this variation might relate to tissue- and cell-specific diseases.
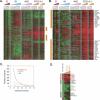
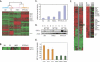
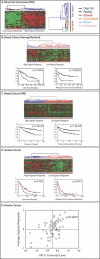
Methods and findings: We analyzed the temporal changes in global transcript levels in response to hypoxia in primary renal proximal tubule epithelial cells, breast epithelial cells, smooth muscle cells, and endothelial cells with DNA microarrays. The extent of the transcriptional response to hypoxia was greatest in the renal tubule cells. This heightened response was associated with a uniquely high level of HIF-1alpha RNA in renal cells, and it could be diminished by reducing HIF-1alpha expression via RNA interference. A gene-expression signature of the hypoxia response, derived from our studies of cultured mammary and renal tubular epithelial cells, showed coordinated variation in several human cancers, and was a strong predictor of clinical outcomes in breast and ovarian cancers. In an analysis of a large, published gene-expression dataset from breast cancers, we found that the prognostic information in the hypoxia signature was virtually independent of that provided by the previously reported wound signature and more predictive of outcomes than any of the clinical parameters in current use.
Conclusions: The transcriptional response to hypoxia varies among human cells. Some of this variation is traceable to variation in expression of the HIF1A gene. A gene-expression signature of the cellular response to hypoxia is associated with a significantly poorer prognosis in breast and ovarian cancer.
Conflict of interest statement
Figures


Comment in
-
Differences in mitotic activity may explain observed differences in HIF-1alpha response in cancers and stroma.PLoS Med. 2006 Aug;3(8):e370. doi: 10.1371/journal.pmed.0030370. PLoS Med. 2006. PMID: 16942404 Free PMC article. No abstract available.
References
-
- Hewitson KS, Schofield CJ. The HIF pathway as a therapeutic target. Drug Discov Today. 2004;9:704–711. - PubMed
-
- Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med. 1996;335:865–875. - PubMed
-
- Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, et al. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995–11998. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

