HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations
- PMID: 16418268
- PMCID: PMC1347991
- DOI: 10.1073/pnas.0508452103
HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations
Abstract
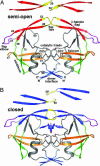
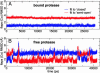
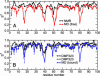
We report unrestrained, all-atom molecular dynamics simulations of HIV-1 protease that sample large conformational changes of the active site flaps. In particular, the unliganded protease undergoes multiple conversions between the "closed" and "semiopen" forms observed in crystal structures of inhibitor-bound and unliganded protease, respectively, including reversal of flap "handedness." Simulations in the presence of a cyclic urea inhibitor yield stable closed flaps. Furthermore, we observe several events in which the flaps of the unliganded protease open to a much greater degree than observed in crystal structures and subsequently return to the semiopen state. Our data strongly support the hypothesis that the unliganded protease predominantly populates the semiopen conformation, with closed and fully open structures being a minor component of the overall ensemble. The results also provide a model for the flap opening and closing that is considered to be essential to enzyme function.
Figures

References
-
- Vondrasek, J. & Wlodawer, A. (2002) Proteins Struct. Funct. Genet. 49, 429–431. - PubMed
-
- Navia, M. A., Fitzgerald, P. M. D., McKeever, B. M., Leu, C. T., Heimbach, J. C., Herber, W. K., Sigal, I. S., Darke, P. L. & Springer, J. P. (1989) Nature 337, 615–620. - PubMed
-
- Wlodawer, A., Miller, M., Jaskolski, M., Sathyanarayana, B. K., Baldwin, E., Weber, I. T., Selk, L. M., Clawson, L., Schneider, J. & Kent, S. B. H. (1989) Science 245, 616–621. - PubMed
-
- Lapatto, R., Blundell, T., Hemmings, A., Overington, J., Wilderspin, A., Wood, S., Merson, J. R., Whittle, P. J., Danley, D. E., Geoghegan, K. F., et al. (1989) Nature 342, 299–302. - PubMed
-
- Nicholson, L. K., Yamazaki, T., Torchia, D. A., Grzesiek, S., Bax, A., Stahl, S. J., Kaufman, J. D., Wingfield, P. T., Lam, P. Y. S., Jadhav, P. K., et al. (1995) Nat. Struct. Biol. 2, 274–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

