ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable
- PMID: 16418286
- PMCID: PMC1348019
- DOI: 10.1073/pnas.0510485103
ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable
Abstract
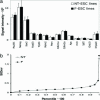
Reproductive cloning is uniformly rejected as a valid technology in humans because of the severely abnormal phenotypes seen in cloned animals. Gene expression aberrations observed in tissues of cloned animals have also raised concerns regarding the therapeutic application of "customized" embryonic stem (ES) cells derived by nuclear transplantation (NT) from a patient's somatic cells. Although previous experiments in mice have demonstrated that the developmental potential of ES cells derived from cloned blastocysts (NT-ES cells) is identical to that of ES cells derived from fertilized blastocysts, a systematic molecular characterization of NT-ES cell lines is lacking. To investigate whether transcriptional aberrations, similar to those observed in tissues of cloned mice, also occur in NT-ES cells, we have compared transcriptional profiles of 10 mouse NT- and fertilization-derived-ES cell lines. We report here that the ES cell lines derived from cloned and fertilized mouse blastocysts are indistinguishable based on their transcriptional profiles, consistent with their normal developmental potential. Our results indicate that, in contrast to embryonic and fetal development of clones, the process of NT-ES cell derivation rigorously selects for those immortal cells that have erased the "epigenetic memory" of the donor nucleus and, thus, become functionally equivalent. Our findings support the notion that ES cell lines derived from cloned or fertilized blastocysts have an identical therapeutic potential.
Figures


References
-
- Munsie, M. J., Michalska, A. E., O'Brien, C. M., Trounson, A. O., Pera, M. F. & Mountford, P. S. (2000) Curr. Biol. 10, 989–992. - PubMed
-
- Wakayama, T., Tabar, V., Rodriguez, I., Perry, A. C., Studer, L. & Mombaerts, P. (2001) Science 292, 740–743. - PubMed
-
- Rideout, W. M., 3rd, Hochedlinger, K., Kyba, M., Daley, G. Q. & Jaenisch, R. (2002) Cell 109, 17–27. - PubMed
-
- Hochedlinger, K. & Jaenisch, R. (2003) N. Engl. J. Med. 349, 275–286. - PubMed
-
- Kohda, T., Inoue, K., Ogonuki, N., Miki, H., Naruse, M., Kaneko-Ishino, T., Ogura, A. & Ishino, F. (2005) Biol. Reprod. 73, 1302–1311. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

